
Chiloscyllium punctatum (Brownbanded bambooshark) (Hemiscyllium punctatum)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Chondrichthyes; Elasmobranchii; Selachii; Galeomorphii; Galeoidea; Orectolobiformes; Hemiscylliidae; Chiloscyllium
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
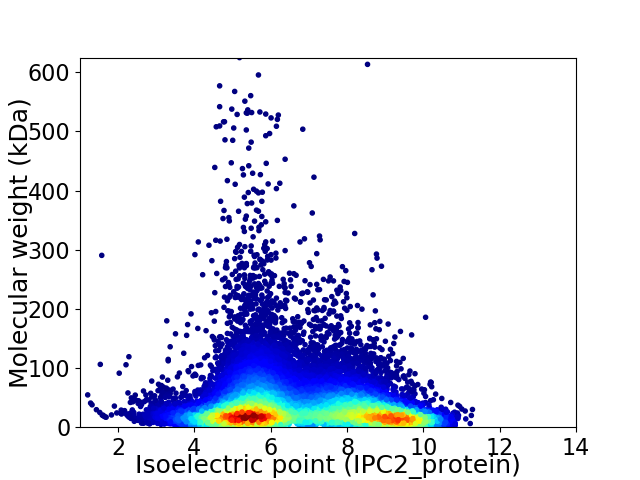
Virtual 2D-PAGE plot for 33497 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A401TEQ2|A0A401TEQ2_CHIPU Uncharacterized protein OS=Chiloscyllium punctatum OX=137246 GN=chiPu_0025122 PE=4 SV=1
LL1 pKa = 8.01VSGSDD6 pKa = 3.54DD7 pKa = 3.69HH8 pKa = 7.82SVSGSDD14 pKa = 2.9NHH16 pKa = 6.13TVSGSDD22 pKa = 3.57DD23 pKa = 3.75QPVSGSDD30 pKa = 3.32DD31 pKa = 3.7HH32 pKa = 7.33AVSGSDD38 pKa = 3.08VHH40 pKa = 6.1TVSGSDD46 pKa = 3.27DD47 pKa = 3.74HH48 pKa = 7.38AVSEE52 pKa = 4.58SDD54 pKa = 3.68VPPVSGSYY62 pKa = 10.36DD63 pKa = 3.56HH64 pKa = 7.03LVSGCDD70 pKa = 3.37DD71 pKa = 4.04HH72 pKa = 7.07PVSVSDD78 pKa = 4.0DD79 pKa = 3.79CLVSGNDD86 pKa = 3.22DD87 pKa = 3.72HH88 pKa = 7.96AGSRR92 pKa = 11.84SDD94 pKa = 3.2VHH96 pKa = 7.17AVSGSDD102 pKa = 3.54DD103 pKa = 3.66QSVSGSDD110 pKa = 3.65DD111 pKa = 3.75DD112 pKa = 4.96AVSGSDD118 pKa = 3.1VHH120 pKa = 7.3AVSGSDD126 pKa = 3.81DD127 pKa = 3.95EE128 pKa = 4.74PVSGSDD134 pKa = 3.28DD135 pKa = 3.65HH136 pKa = 7.23AVSGISEE143 pKa = 4.42SDD145 pKa = 3.6DD146 pKa = 3.55QPVSGGDD153 pKa = 3.55DD154 pKa = 3.47NAVSGIDD161 pKa = 3.68DD162 pKa = 3.89QPVSGSDD169 pKa = 3.58DD170 pKa = 3.7QPDD173 pKa = 3.98SVSDD177 pKa = 3.83DD178 pKa = 3.88SLVSGSDD185 pKa = 3.44DD186 pKa = 3.17QAVFGNDD193 pKa = 2.85DD194 pKa = 3.8HH195 pKa = 7.26AVSGSDD201 pKa = 3.27DD202 pKa = 3.78HH203 pKa = 7.4AVSGSDD209 pKa = 3.38DD210 pKa = 3.83HH211 pKa = 7.95ADD213 pKa = 3.72SGSDD217 pKa = 3.34DD218 pKa = 3.88HH219 pKa = 7.42AVSGNDD225 pKa = 3.23DD226 pKa = 4.15HH227 pKa = 7.22PVSGCDD233 pKa = 3.19DD234 pKa = 3.98HH235 pKa = 8.87SVSEE239 pKa = 4.47SDD241 pKa = 5.01DD242 pKa = 4.15LPLSGSDD249 pKa = 3.49DD250 pKa = 4.07HH251 pKa = 6.99PVSGSDD257 pKa = 3.37DD258 pKa = 3.48HH259 pKa = 7.74SLSGRR264 pKa = 11.84HH265 pKa = 5.7DD266 pKa = 3.6HH267 pKa = 6.08TVSGSGDD274 pKa = 3.63HH275 pKa = 6.65PVPGSDD281 pKa = 3.34DD282 pKa = 4.11HH283 pKa = 7.27PVLGCDD289 pKa = 3.42DD290 pKa = 4.4HH291 pKa = 7.09PVPEE295 pKa = 5.41SDD297 pKa = 3.49DD298 pKa = 3.89HH299 pKa = 6.85SVSVRR304 pKa = 11.84DD305 pKa = 4.02DD306 pKa = 3.69QPVSGSDD313 pKa = 3.52DD314 pKa = 3.73QPVSGSDD321 pKa = 2.78HH322 pKa = 6.07HH323 pKa = 6.66TFSGSEE329 pKa = 4.16INWSLGVMISQFLGVMLMQLLGVMISQFLGVMITQCLGVMISQFLL374 pKa = 3.45
LL1 pKa = 8.01VSGSDD6 pKa = 3.54DD7 pKa = 3.69HH8 pKa = 7.82SVSGSDD14 pKa = 2.9NHH16 pKa = 6.13TVSGSDD22 pKa = 3.57DD23 pKa = 3.75QPVSGSDD30 pKa = 3.32DD31 pKa = 3.7HH32 pKa = 7.33AVSGSDD38 pKa = 3.08VHH40 pKa = 6.1TVSGSDD46 pKa = 3.27DD47 pKa = 3.74HH48 pKa = 7.38AVSEE52 pKa = 4.58SDD54 pKa = 3.68VPPVSGSYY62 pKa = 10.36DD63 pKa = 3.56HH64 pKa = 7.03LVSGCDD70 pKa = 3.37DD71 pKa = 4.04HH72 pKa = 7.07PVSVSDD78 pKa = 4.0DD79 pKa = 3.79CLVSGNDD86 pKa = 3.22DD87 pKa = 3.72HH88 pKa = 7.96AGSRR92 pKa = 11.84SDD94 pKa = 3.2VHH96 pKa = 7.17AVSGSDD102 pKa = 3.54DD103 pKa = 3.66QSVSGSDD110 pKa = 3.65DD111 pKa = 3.75DD112 pKa = 4.96AVSGSDD118 pKa = 3.1VHH120 pKa = 7.3AVSGSDD126 pKa = 3.81DD127 pKa = 3.95EE128 pKa = 4.74PVSGSDD134 pKa = 3.28DD135 pKa = 3.65HH136 pKa = 7.23AVSGISEE143 pKa = 4.42SDD145 pKa = 3.6DD146 pKa = 3.55QPVSGGDD153 pKa = 3.55DD154 pKa = 3.47NAVSGIDD161 pKa = 3.68DD162 pKa = 3.89QPVSGSDD169 pKa = 3.58DD170 pKa = 3.7QPDD173 pKa = 3.98SVSDD177 pKa = 3.83DD178 pKa = 3.88SLVSGSDD185 pKa = 3.44DD186 pKa = 3.17QAVFGNDD193 pKa = 2.85DD194 pKa = 3.8HH195 pKa = 7.26AVSGSDD201 pKa = 3.27DD202 pKa = 3.78HH203 pKa = 7.4AVSGSDD209 pKa = 3.38DD210 pKa = 3.83HH211 pKa = 7.95ADD213 pKa = 3.72SGSDD217 pKa = 3.34DD218 pKa = 3.88HH219 pKa = 7.42AVSGNDD225 pKa = 3.23DD226 pKa = 4.15HH227 pKa = 7.22PVSGCDD233 pKa = 3.19DD234 pKa = 3.98HH235 pKa = 8.87SVSEE239 pKa = 4.47SDD241 pKa = 5.01DD242 pKa = 4.15LPLSGSDD249 pKa = 3.49DD250 pKa = 4.07HH251 pKa = 6.99PVSGSDD257 pKa = 3.37DD258 pKa = 3.48HH259 pKa = 7.74SLSGRR264 pKa = 11.84HH265 pKa = 5.7DD266 pKa = 3.6HH267 pKa = 6.08TVSGSGDD274 pKa = 3.63HH275 pKa = 6.65PVPGSDD281 pKa = 3.34DD282 pKa = 4.11HH283 pKa = 7.27PVLGCDD289 pKa = 3.42DD290 pKa = 4.4HH291 pKa = 7.09PVPEE295 pKa = 5.41SDD297 pKa = 3.49DD298 pKa = 3.89HH299 pKa = 6.85SVSVRR304 pKa = 11.84DD305 pKa = 4.02DD306 pKa = 3.69QPVSGSDD313 pKa = 3.52DD314 pKa = 3.73QPVSGSDD321 pKa = 2.78HH322 pKa = 6.07HH323 pKa = 6.66TFSGSEE329 pKa = 4.16INWSLGVMISQFLGVMLMQLLGVMISQFLGVMITQCLGVMISQFLL374 pKa = 3.45
Molecular weight: 37.93 kDa
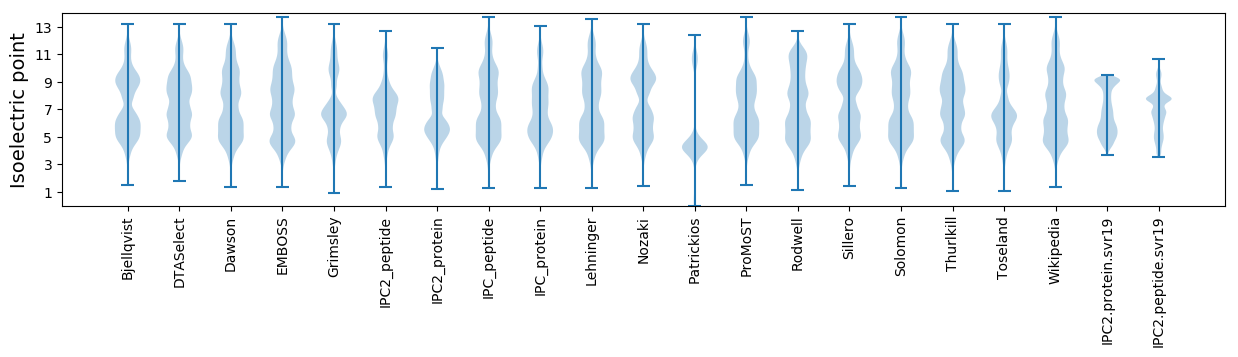
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A401RMS1|A0A401RMS1_CHIPU Uncharacterized protein OS=Chiloscyllium punctatum OX=137246 GN=chiPu_0021063 PE=4 SV=1
MM1 pKa = 7.78ASLLLAPNTGRR12 pKa = 11.84MASLLLAPNTGRR24 pKa = 11.84MASLLLAPNAEE35 pKa = 3.99RR36 pKa = 11.84MALLLLAPNAGRR48 pKa = 11.84MALLLLAPNAGRR60 pKa = 11.84MASLLPAPNARR71 pKa = 11.84RR72 pKa = 11.84MDD74 pKa = 4.06SLLLAPNARR83 pKa = 11.84RR84 pKa = 11.84MASLLLALNARR95 pKa = 11.84KK96 pKa = 8.26MVSLLLAPHH105 pKa = 6.96ARR107 pKa = 11.84RR108 pKa = 11.84MASLLLVTQARR119 pKa = 11.84RR120 pKa = 11.84MASLLVAPNARR131 pKa = 11.84RR132 pKa = 11.84MVSLLLAPHH141 pKa = 6.96ARR143 pKa = 11.84RR144 pKa = 11.84MASLLLAPNARR155 pKa = 11.84RR156 pKa = 11.84MASLLLAPHH165 pKa = 6.64FRR167 pKa = 11.84RR168 pKa = 11.84MATLLLAPNARR179 pKa = 11.84RR180 pKa = 11.84MASLLLAPHH189 pKa = 7.03ARR191 pKa = 11.84RR192 pKa = 11.84MASLLLASQVRR203 pKa = 11.84RR204 pKa = 11.84MASLLLAPNARR215 pKa = 11.84RR216 pKa = 11.84MVLMLLSPHH225 pKa = 6.29ARR227 pKa = 11.84RR228 pKa = 11.84MALLLLAPNAGRR240 pKa = 11.84MASLLLAPNAKK251 pKa = 9.81RR252 pKa = 11.84MASLLPALNARR263 pKa = 11.84RR264 pKa = 11.84MALLLLAPNAGRR276 pKa = 11.84MAA278 pKa = 4.48
MM1 pKa = 7.78ASLLLAPNTGRR12 pKa = 11.84MASLLLAPNTGRR24 pKa = 11.84MASLLLAPNAEE35 pKa = 3.99RR36 pKa = 11.84MALLLLAPNAGRR48 pKa = 11.84MALLLLAPNAGRR60 pKa = 11.84MASLLPAPNARR71 pKa = 11.84RR72 pKa = 11.84MDD74 pKa = 4.06SLLLAPNARR83 pKa = 11.84RR84 pKa = 11.84MASLLLALNARR95 pKa = 11.84KK96 pKa = 8.26MVSLLLAPHH105 pKa = 6.96ARR107 pKa = 11.84RR108 pKa = 11.84MASLLLVTQARR119 pKa = 11.84RR120 pKa = 11.84MASLLVAPNARR131 pKa = 11.84RR132 pKa = 11.84MVSLLLAPHH141 pKa = 6.96ARR143 pKa = 11.84RR144 pKa = 11.84MASLLLAPNARR155 pKa = 11.84RR156 pKa = 11.84MASLLLAPHH165 pKa = 6.64FRR167 pKa = 11.84RR168 pKa = 11.84MATLLLAPNARR179 pKa = 11.84RR180 pKa = 11.84MASLLLAPHH189 pKa = 7.03ARR191 pKa = 11.84RR192 pKa = 11.84MASLLLASQVRR203 pKa = 11.84RR204 pKa = 11.84MASLLLAPNARR215 pKa = 11.84RR216 pKa = 11.84MVLMLLSPHH225 pKa = 6.29ARR227 pKa = 11.84RR228 pKa = 11.84MALLLLAPNAGRR240 pKa = 11.84MASLLLAPNAKK251 pKa = 9.81RR252 pKa = 11.84MASLLPALNARR263 pKa = 11.84RR264 pKa = 11.84MALLLLAPNAGRR276 pKa = 11.84MAA278 pKa = 4.48
Molecular weight: 29.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12024568 |
30 |
8400 |
359.0 |
40.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.572 ± 0.017 | 2.23 ± 0.013 |
5.295 ± 0.022 | 7.19 ± 0.025 |
3.425 ± 0.011 | 6.543 ± 0.025 |
2.517 ± 0.008 | 4.873 ± 0.013 |
5.751 ± 0.018 | 9.184 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.309 ± 0.007 | 4.122 ± 0.012 |
5.247 ± 0.02 | 4.693 ± 0.017 |
5.961 ± 0.021 | 8.21 ± 0.025 |
5.591 ± 0.018 | 6.422 ± 0.017 |
1.203 ± 0.007 | 2.663 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
