
Actinokineospora terrae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Actinokineospora
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
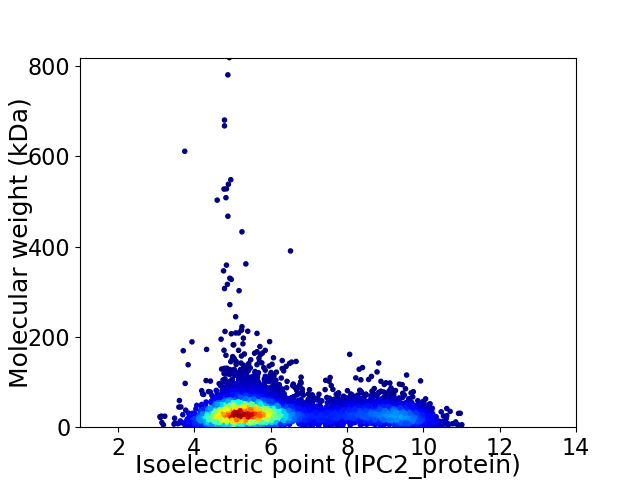
Virtual 2D-PAGE plot for 6615 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9XPG1|A0A1H9XPG1_9PSEU Uncharacterized protein OS=Actinokineospora terrae OX=155974 GN=SAMN04487818_11871 PE=4 SV=1
MM1 pKa = 8.02PIGTTRR7 pKa = 11.84PRR9 pKa = 11.84AAIGRR14 pKa = 11.84RR15 pKa = 11.84VRR17 pKa = 11.84TLTCAVIMAAALPVVVGPPAQATFPGINGKK47 pKa = 9.65IYY49 pKa = 10.24FDD51 pKa = 3.83QDD53 pKa = 3.61DD54 pKa = 4.19EE55 pKa = 5.14VYY57 pKa = 10.73SINPDD62 pKa = 3.2GTGLTQLTDD71 pKa = 3.5SDD73 pKa = 5.12DD74 pKa = 3.66LSKK77 pKa = 11.09DD78 pKa = 3.2PRR80 pKa = 11.84VNADD84 pKa = 2.97GSKK87 pKa = 10.25VAFLRR92 pKa = 11.84YY93 pKa = 9.65SFSQGSNQVWVMNPDD108 pKa = 3.13GTGATQLTSAISADD122 pKa = 3.8SGSLAWSPAGDD133 pKa = 3.44KK134 pKa = 10.62LVYY137 pKa = 10.71SPDD140 pKa = 3.53LLTVIDD146 pKa = 5.3ADD148 pKa = 3.87GTDD151 pKa = 3.82PFSLGVSGGAPTWSPDD167 pKa = 3.03GTRR170 pKa = 11.84IAYY173 pKa = 6.56TASGGIRR180 pKa = 11.84AINPDD185 pKa = 3.0GTGQTTIKK193 pKa = 10.29NRR195 pKa = 11.84PSPPEE200 pKa = 3.96FLSGPSWSPDD210 pKa = 2.78GTKK213 pKa = 9.82IAASTYY219 pKa = 10.8DD220 pKa = 3.37FSTGQGGVFTINADD234 pKa = 3.21GTGYY238 pKa = 9.63TNISGTNTDD247 pKa = 3.93DD248 pKa = 4.51GAAVWSPDD256 pKa = 2.72GTKK259 pKa = 9.98ISFHH263 pKa = 7.41DD264 pKa = 4.03GTGLAVMNTNGTGRR278 pKa = 11.84TALHH282 pKa = 6.63SGFLNNGDD290 pKa = 3.52WAAVPAGADD299 pKa = 3.29VSVSVTDD306 pKa = 3.47SADD309 pKa = 3.54PVSLGGSPYY318 pKa = 9.73TYY320 pKa = 9.65TVSVANAGPAGATGVSVSTTLSGANRR346 pKa = 11.84TINSATSSQGSCTVSAPTVSCALGAVAASGSATVTISVTPSATGTITATSTVSATEE402 pKa = 3.87TDD404 pKa = 3.74PLSGNNTAAQSTTVNNALGCAITGTPGNDD433 pKa = 3.15TLNGGNGNDD442 pKa = 4.02VICGLGGNDD451 pKa = 4.65VINGGNNNDD460 pKa = 4.12TIYY463 pKa = 11.04AGAGDD468 pKa = 3.97DD469 pKa = 4.53TIDD472 pKa = 4.84GGNSDD477 pKa = 4.34DD478 pKa = 4.78TIHH481 pKa = 7.1AGDD484 pKa = 4.28GDD486 pKa = 4.28DD487 pKa = 5.12VIGGGNSADD496 pKa = 3.74YY497 pKa = 10.93LYY499 pKa = 11.45GEE501 pKa = 5.24AGNDD505 pKa = 3.34TNYY508 pKa = 11.11GEE510 pKa = 4.44TFLGSLLYY518 pKa = 11.11LFDD521 pKa = 4.45NGNDD525 pKa = 3.92HH526 pKa = 7.54IYY528 pKa = 10.76GGPGNDD534 pKa = 5.36DD535 pKa = 4.48LDD537 pKa = 4.22GQNGNDD543 pKa = 4.09TIVDD547 pKa = 3.62HH548 pKa = 7.68DD549 pKa = 4.21GTDD552 pKa = 3.02IMSGSNGNDD561 pKa = 3.46TIDD564 pKa = 3.46VLDD567 pKa = 4.24GVGGDD572 pKa = 3.74TANGGLGSDD581 pKa = 3.74TCAVDD586 pKa = 4.42GGDD589 pKa = 3.59TATGCC594 pKa = 4.36
MM1 pKa = 8.02PIGTTRR7 pKa = 11.84PRR9 pKa = 11.84AAIGRR14 pKa = 11.84RR15 pKa = 11.84VRR17 pKa = 11.84TLTCAVIMAAALPVVVGPPAQATFPGINGKK47 pKa = 9.65IYY49 pKa = 10.24FDD51 pKa = 3.83QDD53 pKa = 3.61DD54 pKa = 4.19EE55 pKa = 5.14VYY57 pKa = 10.73SINPDD62 pKa = 3.2GTGLTQLTDD71 pKa = 3.5SDD73 pKa = 5.12DD74 pKa = 3.66LSKK77 pKa = 11.09DD78 pKa = 3.2PRR80 pKa = 11.84VNADD84 pKa = 2.97GSKK87 pKa = 10.25VAFLRR92 pKa = 11.84YY93 pKa = 9.65SFSQGSNQVWVMNPDD108 pKa = 3.13GTGATQLTSAISADD122 pKa = 3.8SGSLAWSPAGDD133 pKa = 3.44KK134 pKa = 10.62LVYY137 pKa = 10.71SPDD140 pKa = 3.53LLTVIDD146 pKa = 5.3ADD148 pKa = 3.87GTDD151 pKa = 3.82PFSLGVSGGAPTWSPDD167 pKa = 3.03GTRR170 pKa = 11.84IAYY173 pKa = 6.56TASGGIRR180 pKa = 11.84AINPDD185 pKa = 3.0GTGQTTIKK193 pKa = 10.29NRR195 pKa = 11.84PSPPEE200 pKa = 3.96FLSGPSWSPDD210 pKa = 2.78GTKK213 pKa = 9.82IAASTYY219 pKa = 10.8DD220 pKa = 3.37FSTGQGGVFTINADD234 pKa = 3.21GTGYY238 pKa = 9.63TNISGTNTDD247 pKa = 3.93DD248 pKa = 4.51GAAVWSPDD256 pKa = 2.72GTKK259 pKa = 9.98ISFHH263 pKa = 7.41DD264 pKa = 4.03GTGLAVMNTNGTGRR278 pKa = 11.84TALHH282 pKa = 6.63SGFLNNGDD290 pKa = 3.52WAAVPAGADD299 pKa = 3.29VSVSVTDD306 pKa = 3.47SADD309 pKa = 3.54PVSLGGSPYY318 pKa = 9.73TYY320 pKa = 9.65TVSVANAGPAGATGVSVSTTLSGANRR346 pKa = 11.84TINSATSSQGSCTVSAPTVSCALGAVAASGSATVTISVTPSATGTITATSTVSATEE402 pKa = 3.87TDD404 pKa = 3.74PLSGNNTAAQSTTVNNALGCAITGTPGNDD433 pKa = 3.15TLNGGNGNDD442 pKa = 4.02VICGLGGNDD451 pKa = 4.65VINGGNNNDD460 pKa = 4.12TIYY463 pKa = 11.04AGAGDD468 pKa = 3.97DD469 pKa = 4.53TIDD472 pKa = 4.84GGNSDD477 pKa = 4.34DD478 pKa = 4.78TIHH481 pKa = 7.1AGDD484 pKa = 4.28GDD486 pKa = 4.28DD487 pKa = 5.12VIGGGNSADD496 pKa = 3.74YY497 pKa = 10.93LYY499 pKa = 11.45GEE501 pKa = 5.24AGNDD505 pKa = 3.34TNYY508 pKa = 11.11GEE510 pKa = 4.44TFLGSLLYY518 pKa = 11.11LFDD521 pKa = 4.45NGNDD525 pKa = 3.92HH526 pKa = 7.54IYY528 pKa = 10.76GGPGNDD534 pKa = 5.36DD535 pKa = 4.48LDD537 pKa = 4.22GQNGNDD543 pKa = 4.09TIVDD547 pKa = 3.62HH548 pKa = 7.68DD549 pKa = 4.21GTDD552 pKa = 3.02IMSGSNGNDD561 pKa = 3.46TIDD564 pKa = 3.46VLDD567 pKa = 4.24GVGGDD572 pKa = 3.74TANGGLGSDD581 pKa = 3.74TCAVDD586 pKa = 4.42GGDD589 pKa = 3.59TATGCC594 pKa = 4.36
Molecular weight: 59.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9XRS5|A0A1H9XRS5_9PSEU Uncharacterized protein OS=Actinokineospora terrae OX=155974 GN=SAMN04487818_12310 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84DD41 pKa = 3.0KK42 pKa = 11.37LSAA45 pKa = 3.85
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AIIAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84DD41 pKa = 3.0KK42 pKa = 11.37LSAA45 pKa = 3.85
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2246422 |
39 |
7858 |
339.6 |
36.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.6 ± 0.044 | 0.754 ± 0.009 |
6.248 ± 0.024 | 5.094 ± 0.029 |
2.721 ± 0.018 | 9.102 ± 0.029 |
2.251 ± 0.015 | 3.249 ± 0.018 |
1.882 ± 0.024 | 10.461 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.516 ± 0.014 | 1.84 ± 0.018 |
6.007 ± 0.031 | 2.696 ± 0.019 |
7.814 ± 0.04 | 5.13 ± 0.026 |
6.689 ± 0.043 | 9.525 ± 0.034 |
1.52 ± 0.014 | 1.902 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
