
Thiorhodospira sibirica ATCC 700588
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; Thiorhodospira; Thiorhodospira sibirica
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
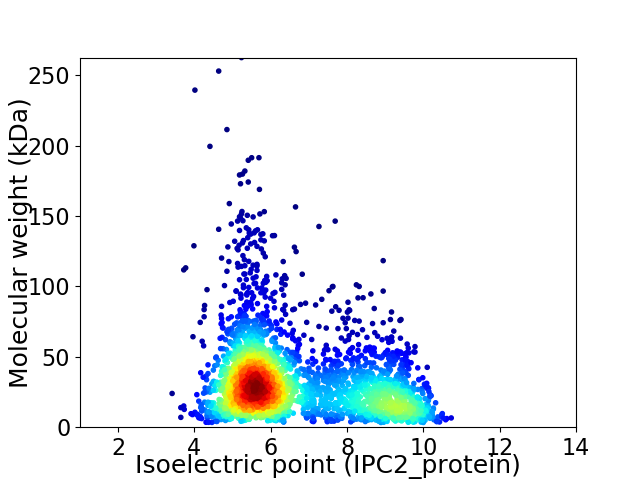
Virtual 2D-PAGE plot for 2933 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G4E8T0|G4E8T0_9GAMM UvrABC system protein C OS=Thiorhodospira sibirica ATCC 700588 OX=765914 GN=uvrC PE=3 SV=1
MM1 pKa = 7.14LTIDD5 pKa = 4.4PGLVCWLIEE14 pKa = 4.46RR15 pKa = 11.84IQEE18 pKa = 3.53FHH20 pKa = 6.46AQEE23 pKa = 4.06DD24 pKa = 4.5VAIHH28 pKa = 6.04EE29 pKa = 4.64EE30 pKa = 4.33PEE32 pKa = 4.09IPHH35 pKa = 7.23DD36 pKa = 3.91DD37 pKa = 3.27WGLAGMASVAEE48 pKa = 4.3YY49 pKa = 10.91ADD51 pKa = 3.9DD52 pKa = 4.3PAYY55 pKa = 10.63QEE57 pKa = 3.93IKK59 pKa = 11.1SNIDD63 pKa = 3.15EE64 pKa = 5.06LEE66 pKa = 4.48PEE68 pKa = 4.14QQVCLVALMWMGRR81 pKa = 11.84GDD83 pKa = 3.76FDD85 pKa = 5.66DD86 pKa = 6.0NEE88 pKa = 4.15WQLAFAEE95 pKa = 4.99AKK97 pKa = 10.65SNWTSHH103 pKa = 3.99TAEE106 pKa = 4.24YY107 pKa = 10.48LIATPLVSDD116 pKa = 4.12YY117 pKa = 11.0LAEE120 pKa = 4.44GLSMMGYY127 pKa = 9.65SCDD130 pKa = 5.33DD131 pKa = 3.31EE132 pKa = 5.02DD133 pKa = 4.49
MM1 pKa = 7.14LTIDD5 pKa = 4.4PGLVCWLIEE14 pKa = 4.46RR15 pKa = 11.84IQEE18 pKa = 3.53FHH20 pKa = 6.46AQEE23 pKa = 4.06DD24 pKa = 4.5VAIHH28 pKa = 6.04EE29 pKa = 4.64EE30 pKa = 4.33PEE32 pKa = 4.09IPHH35 pKa = 7.23DD36 pKa = 3.91DD37 pKa = 3.27WGLAGMASVAEE48 pKa = 4.3YY49 pKa = 10.91ADD51 pKa = 3.9DD52 pKa = 4.3PAYY55 pKa = 10.63QEE57 pKa = 3.93IKK59 pKa = 11.1SNIDD63 pKa = 3.15EE64 pKa = 5.06LEE66 pKa = 4.48PEE68 pKa = 4.14QQVCLVALMWMGRR81 pKa = 11.84GDD83 pKa = 3.76FDD85 pKa = 5.66DD86 pKa = 6.0NEE88 pKa = 4.15WQLAFAEE95 pKa = 4.99AKK97 pKa = 10.65SNWTSHH103 pKa = 3.99TAEE106 pKa = 4.24YY107 pKa = 10.48LIATPLVSDD116 pKa = 4.12YY117 pKa = 11.0LAEE120 pKa = 4.44GLSMMGYY127 pKa = 9.65SCDD130 pKa = 5.33DD131 pKa = 3.31EE132 pKa = 5.02DD133 pKa = 4.49
Molecular weight: 15.06 kDa
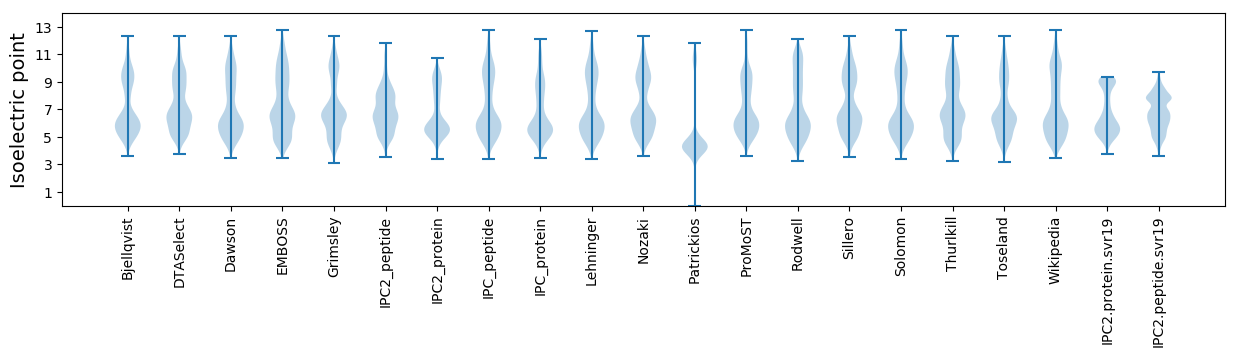
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G4E913|G4E913_9GAMM Adenine specific DNA methyltransferase (Fragment) OS=Thiorhodospira sibirica ATCC 700588 OX=765914 GN=ThisiDRAFT_2792 PE=4 SV=1
MM1 pKa = 6.96TKK3 pKa = 9.87EE4 pKa = 4.66HH5 pKa = 6.25ITMSHH10 pKa = 6.3KK11 pKa = 10.74EE12 pKa = 3.62IDD14 pKa = 3.93RR15 pKa = 11.84LEE17 pKa = 4.47MIQAVANKK25 pKa = 9.68HH26 pKa = 5.35LRR28 pKa = 11.84QAEE31 pKa = 3.9AAQRR35 pKa = 11.84LGLSVRR41 pKa = 11.84QVKK44 pKa = 10.25RR45 pKa = 11.84LVRR48 pKa = 11.84RR49 pKa = 11.84YY50 pKa = 9.34RR51 pKa = 11.84EE52 pKa = 3.9HH53 pKa = 6.84GATGLRR59 pKa = 11.84SGHH62 pKa = 6.46RR63 pKa = 11.84GRR65 pKa = 11.84RR66 pKa = 11.84PNNAIAEE73 pKa = 4.53TVRR76 pKa = 11.84QEE78 pKa = 3.75VLALVKK84 pKa = 8.87THH86 pKa = 5.74YY87 pKa = 9.91TDD89 pKa = 5.04FGPTLACEE97 pKa = 4.36KK98 pKa = 9.91LAEE101 pKa = 4.18RR102 pKa = 11.84HH103 pKa = 4.8GHH105 pKa = 5.44HH106 pKa = 6.84LSVEE110 pKa = 4.34TLRR113 pKa = 11.84QWMVADD119 pKa = 4.87GLWQPKK125 pKa = 7.08QRR127 pKa = 11.84KK128 pKa = 5.46QARR131 pKa = 11.84IHH133 pKa = 4.62QRR135 pKa = 11.84RR136 pKa = 11.84PRR138 pKa = 11.84RR139 pKa = 11.84PCLGEE144 pKa = 4.74LIQIDD149 pKa = 4.8GSPHH153 pKa = 6.22DD154 pKa = 3.8WFEE157 pKa = 4.06VRR159 pKa = 11.84GPRR162 pKa = 11.84CTLMVFIDD170 pKa = 4.27DD171 pKa = 3.55ATGRR175 pKa = 11.84LMALRR180 pKa = 11.84FVPAEE185 pKa = 3.83TTQAYY190 pKa = 8.75MEE192 pKa = 4.44TLQQYY197 pKa = 10.36LDD199 pKa = 3.33QHH201 pKa = 5.98GRR203 pKa = 11.84PVALYY208 pKa = 9.85SDD210 pKa = 3.34KK211 pKa = 11.26HH212 pKa = 7.26SIFRR216 pKa = 11.84VNHH219 pKa = 6.38PEE221 pKa = 3.61HH222 pKa = 7.23DD223 pKa = 4.05GEE225 pKa = 4.18LTQFSRR231 pKa = 11.84ALKK234 pKa = 9.01TLDD237 pKa = 3.08IAAIHH242 pKa = 6.74ANTPQAKK249 pKa = 9.79GRR251 pKa = 11.84VEE253 pKa = 3.95RR254 pKa = 11.84ANQTLQDD261 pKa = 3.78RR262 pKa = 11.84LVKK265 pKa = 9.89EE266 pKa = 3.66LRR268 pKa = 11.84LRR270 pKa = 11.84EE271 pKa = 3.95ISDD274 pKa = 3.05IDD276 pKa = 3.47AANAFLPTFMADD288 pKa = 3.3FNARR292 pKa = 11.84FAVEE296 pKa = 4.06PQSPPGLPTDD306 pKa = 4.29RR307 pKa = 11.84CCTAPRR313 pKa = 11.84SRR315 pKa = 11.84PSSFACTIPVSSPRR329 pKa = 11.84TFASSSRR336 pKa = 11.84TGISASGTGARR347 pKa = 11.84ATACGVPPSPSARR360 pKa = 11.84PSTAPSPSCTRR371 pKa = 11.84GTSCLTVCWLRR382 pKa = 11.84ANRR385 pKa = 11.84PHH387 pKa = 6.66RR388 pKa = 11.84WMMKK392 pKa = 9.76KK393 pKa = 10.47VSTTASTRR401 pKa = 11.84PRR403 pKa = 11.84QNRR406 pKa = 11.84PPSQPTNRR414 pKa = 11.84RR415 pKa = 11.84RR416 pKa = 11.84TTLGEE421 pKa = 4.0VGATSLRR428 pKa = 11.84HH429 pKa = 4.66QPKK432 pKa = 8.6HH433 pKa = 6.35HH434 pKa = 5.98NHH436 pKa = 6.24AFYY439 pKa = 10.8GGHH442 pKa = 6.29FCFAQKK448 pKa = 9.38GTFLNWVDD456 pKa = 3.81SRR458 pKa = 11.84NGSPVV463 pKa = 2.73
MM1 pKa = 6.96TKK3 pKa = 9.87EE4 pKa = 4.66HH5 pKa = 6.25ITMSHH10 pKa = 6.3KK11 pKa = 10.74EE12 pKa = 3.62IDD14 pKa = 3.93RR15 pKa = 11.84LEE17 pKa = 4.47MIQAVANKK25 pKa = 9.68HH26 pKa = 5.35LRR28 pKa = 11.84QAEE31 pKa = 3.9AAQRR35 pKa = 11.84LGLSVRR41 pKa = 11.84QVKK44 pKa = 10.25RR45 pKa = 11.84LVRR48 pKa = 11.84RR49 pKa = 11.84YY50 pKa = 9.34RR51 pKa = 11.84EE52 pKa = 3.9HH53 pKa = 6.84GATGLRR59 pKa = 11.84SGHH62 pKa = 6.46RR63 pKa = 11.84GRR65 pKa = 11.84RR66 pKa = 11.84PNNAIAEE73 pKa = 4.53TVRR76 pKa = 11.84QEE78 pKa = 3.75VLALVKK84 pKa = 8.87THH86 pKa = 5.74YY87 pKa = 9.91TDD89 pKa = 5.04FGPTLACEE97 pKa = 4.36KK98 pKa = 9.91LAEE101 pKa = 4.18RR102 pKa = 11.84HH103 pKa = 4.8GHH105 pKa = 5.44HH106 pKa = 6.84LSVEE110 pKa = 4.34TLRR113 pKa = 11.84QWMVADD119 pKa = 4.87GLWQPKK125 pKa = 7.08QRR127 pKa = 11.84KK128 pKa = 5.46QARR131 pKa = 11.84IHH133 pKa = 4.62QRR135 pKa = 11.84RR136 pKa = 11.84PRR138 pKa = 11.84RR139 pKa = 11.84PCLGEE144 pKa = 4.74LIQIDD149 pKa = 4.8GSPHH153 pKa = 6.22DD154 pKa = 3.8WFEE157 pKa = 4.06VRR159 pKa = 11.84GPRR162 pKa = 11.84CTLMVFIDD170 pKa = 4.27DD171 pKa = 3.55ATGRR175 pKa = 11.84LMALRR180 pKa = 11.84FVPAEE185 pKa = 3.83TTQAYY190 pKa = 8.75MEE192 pKa = 4.44TLQQYY197 pKa = 10.36LDD199 pKa = 3.33QHH201 pKa = 5.98GRR203 pKa = 11.84PVALYY208 pKa = 9.85SDD210 pKa = 3.34KK211 pKa = 11.26HH212 pKa = 7.26SIFRR216 pKa = 11.84VNHH219 pKa = 6.38PEE221 pKa = 3.61HH222 pKa = 7.23DD223 pKa = 4.05GEE225 pKa = 4.18LTQFSRR231 pKa = 11.84ALKK234 pKa = 9.01TLDD237 pKa = 3.08IAAIHH242 pKa = 6.74ANTPQAKK249 pKa = 9.79GRR251 pKa = 11.84VEE253 pKa = 3.95RR254 pKa = 11.84ANQTLQDD261 pKa = 3.78RR262 pKa = 11.84LVKK265 pKa = 9.89EE266 pKa = 3.66LRR268 pKa = 11.84LRR270 pKa = 11.84EE271 pKa = 3.95ISDD274 pKa = 3.05IDD276 pKa = 3.47AANAFLPTFMADD288 pKa = 3.3FNARR292 pKa = 11.84FAVEE296 pKa = 4.06PQSPPGLPTDD306 pKa = 4.29RR307 pKa = 11.84CCTAPRR313 pKa = 11.84SRR315 pKa = 11.84PSSFACTIPVSSPRR329 pKa = 11.84TFASSSRR336 pKa = 11.84TGISASGTGARR347 pKa = 11.84ATACGVPPSPSARR360 pKa = 11.84PSTAPSPSCTRR371 pKa = 11.84GTSCLTVCWLRR382 pKa = 11.84ANRR385 pKa = 11.84PHH387 pKa = 6.66RR388 pKa = 11.84WMMKK392 pKa = 9.76KK393 pKa = 10.47VSTTASTRR401 pKa = 11.84PRR403 pKa = 11.84QNRR406 pKa = 11.84PPSQPTNRR414 pKa = 11.84RR415 pKa = 11.84RR416 pKa = 11.84TTLGEE421 pKa = 4.0VGATSLRR428 pKa = 11.84HH429 pKa = 4.66QPKK432 pKa = 8.6HH433 pKa = 6.35HH434 pKa = 5.98NHH436 pKa = 6.24AFYY439 pKa = 10.8GGHH442 pKa = 6.29FCFAQKK448 pKa = 9.38GTFLNWVDD456 pKa = 3.81SRR458 pKa = 11.84NGSPVV463 pKa = 2.73
Molecular weight: 52.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
923034 |
30 |
2404 |
314.7 |
34.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.825 ± 0.055 | 1.085 ± 0.017 |
5.142 ± 0.035 | 5.877 ± 0.048 |
3.472 ± 0.031 | 7.062 ± 0.043 |
2.952 ± 0.027 | 5.342 ± 0.035 |
3.097 ± 0.041 | 11.671 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.361 ± 0.02 | 2.834 ± 0.036 |
5.096 ± 0.038 | 5.212 ± 0.042 |
6.956 ± 0.045 | 5.293 ± 0.034 |
5.128 ± 0.036 | 6.636 ± 0.037 |
1.333 ± 0.021 | 2.625 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
