
Longilinea arvoryzae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Anaerolineae; Anaerolineales; Anaerolineaceae; Longilinea
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
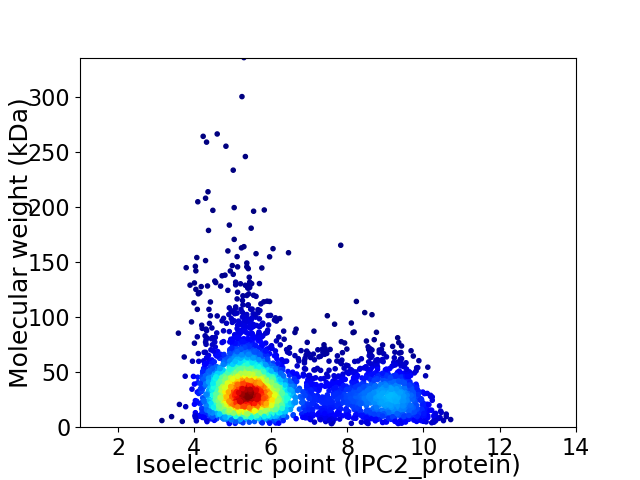
Virtual 2D-PAGE plot for 3871 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S7BDH4|A0A0S7BDH4_9CHLR Molecular chaperone HSP90 family OS=Longilinea arvoryzae OX=360412 GN=LARV_00524 PE=4 SV=1
MM1 pKa = 7.57TNPSNFKK8 pKa = 10.56LINLGLTLIIALGCVFSGITPVQAAAQGLSNTYY41 pKa = 7.83TQSFDD46 pKa = 3.14GLRR49 pKa = 11.84ASGSGAWVNDD59 pKa = 3.11STLPGWYY66 pKa = 9.79ASRR69 pKa = 11.84NQITADD75 pKa = 3.61NGSSGAGGMFSYY87 pKa = 11.04GSTASAEE94 pKa = 4.29RR95 pKa = 11.84AFGALPKK102 pKa = 10.26KK103 pKa = 9.46NTGGTIYY110 pKa = 10.51KK111 pKa = 10.51GLLLQNDD118 pKa = 4.35TLNEE122 pKa = 3.69ITRR125 pKa = 11.84LYY127 pKa = 10.8VAFTGEE133 pKa = 3.65QWRR136 pKa = 11.84NSAGGTQILAFSYY149 pKa = 10.46QVGASAITSLTAGAWTSLTSLDD171 pKa = 3.56FSAPVTSGTAGALNGNLPANQRR193 pKa = 11.84RR194 pKa = 11.84RR195 pKa = 11.84TAQFSVAIPAGSYY208 pKa = 10.82IMLRR212 pKa = 11.84WTDD215 pKa = 3.31ADD217 pKa = 3.77EE218 pKa = 4.22QGADD222 pKa = 3.4HH223 pKa = 7.0GLAIDD228 pKa = 4.15DD229 pKa = 4.54LVVSRR234 pKa = 11.84YY235 pKa = 10.0DD236 pKa = 3.59PPQAAADD243 pKa = 4.3AYY245 pKa = 9.07QTSEE249 pKa = 4.28DD250 pKa = 4.03TPLTIAAPGLLANDD264 pKa = 4.41TAHH267 pKa = 6.69QGQPLSAVLVDD278 pKa = 4.18QPSHH282 pKa = 5.87GAVVLNADD290 pKa = 3.74GSFTYY295 pKa = 10.36TPTADD300 pKa = 3.24WNGSDD305 pKa = 4.08SFTYY309 pKa = 8.32TAKK312 pKa = 10.6EE313 pKa = 3.84AGLASAPAAVTLTISPVNDD332 pKa = 3.6APRR335 pKa = 11.84AAADD339 pKa = 3.76GGTTDD344 pKa = 3.59EE345 pKa = 5.28DD346 pKa = 4.15VPLVQPAPGVLDD358 pKa = 3.83NDD360 pKa = 3.61SDD362 pKa = 4.55ADD364 pKa = 3.83GDD366 pKa = 4.31ALTTALVDD374 pKa = 4.44GPSHH378 pKa = 7.31GSLTLNSDD386 pKa = 3.14GSYY389 pKa = 10.06TYY391 pKa = 10.41TPNADD396 pKa = 3.01WNGTDD401 pKa = 3.05NFTYY405 pKa = 9.74TAFDD409 pKa = 3.63GVVNSSVATVTLTVNPAADD428 pKa = 3.95APRR431 pKa = 11.84TTPDD435 pKa = 3.37SYY437 pKa = 11.88DD438 pKa = 3.29VDD440 pKa = 3.9EE441 pKa = 5.54DD442 pKa = 4.05ALLSIAAPGLLEE454 pKa = 4.77NDD456 pKa = 3.27TDD458 pKa = 4.62ADD460 pKa = 4.09GDD462 pKa = 3.87NLTAVWVSDD471 pKa = 3.78PLHH474 pKa = 6.48GVLALNADD482 pKa = 3.68GSYY485 pKa = 10.03TYY487 pKa = 10.79QPDD490 pKa = 3.97ANWNGDD496 pKa = 3.66DD497 pKa = 3.79SFTYY501 pKa = 9.85QASDD505 pKa = 3.45GFLLSAVEE513 pKa = 4.34AVTLHH518 pKa = 5.27VQPINDD524 pKa = 3.97APTAIEE530 pKa = 4.89DD531 pKa = 4.51GYY533 pKa = 9.96STPEE537 pKa = 3.65DD538 pKa = 3.41TQLNVNAPGVLGNDD552 pKa = 3.52TDD554 pKa = 4.51RR555 pKa = 11.84DD556 pKa = 3.86GDD558 pKa = 3.91GLSAHH563 pKa = 6.89LVTDD567 pKa = 4.62ASHH570 pKa = 7.49GDD572 pKa = 3.34LTLNADD578 pKa = 3.86GSFSYY583 pKa = 10.97QPDD586 pKa = 3.79SDD588 pKa = 3.97WNGDD592 pKa = 3.65DD593 pKa = 3.43SFTYY597 pKa = 10.01AAHH600 pKa = 6.92DD601 pKa = 4.05AEE603 pKa = 4.81STSGTVTVEE612 pKa = 3.78LTVNPVNDD620 pKa = 4.44APTATADD627 pKa = 3.5TYY629 pKa = 11.77SLVEE633 pKa = 4.36NGSLSIPANGVLANDD648 pKa = 3.54IDD650 pKa = 3.97VDD652 pKa = 4.25GDD654 pKa = 3.71VLTAVLDD661 pKa = 3.9SSTSHH666 pKa = 7.03GSLTLNTDD674 pKa = 3.09GSFSYY679 pKa = 10.54TPAADD684 pKa = 3.47WNGSDD689 pKa = 4.28SFTYY693 pKa = 10.04HH694 pKa = 6.85ANDD697 pKa = 3.73GAASSEE703 pKa = 4.49TVTVTLTVDD712 pKa = 3.64PDD714 pKa = 3.1NTAPYY719 pKa = 9.87RR720 pKa = 11.84IAPLPDD726 pKa = 3.15QNHH729 pKa = 7.19PARR732 pKa = 11.84TPYY735 pKa = 10.97SFDD738 pKa = 2.84TSAYY742 pKa = 10.35FGDD745 pKa = 4.4SDD747 pKa = 3.92TGDD750 pKa = 3.34VLTFSAQLVDD760 pKa = 5.05GNPLPPWLSCNTASGVLSGTPPLAAIGVYY789 pKa = 9.64SIRR792 pKa = 11.84VTASDD797 pKa = 3.69GSATVSDD804 pKa = 4.95DD805 pKa = 3.48FDD807 pKa = 4.04LTVEE811 pKa = 4.22DD812 pKa = 3.27NPYY815 pKa = 10.75RR816 pKa = 11.84LFIPMVLRR824 pKa = 4.33
MM1 pKa = 7.57TNPSNFKK8 pKa = 10.56LINLGLTLIIALGCVFSGITPVQAAAQGLSNTYY41 pKa = 7.83TQSFDD46 pKa = 3.14GLRR49 pKa = 11.84ASGSGAWVNDD59 pKa = 3.11STLPGWYY66 pKa = 9.79ASRR69 pKa = 11.84NQITADD75 pKa = 3.61NGSSGAGGMFSYY87 pKa = 11.04GSTASAEE94 pKa = 4.29RR95 pKa = 11.84AFGALPKK102 pKa = 10.26KK103 pKa = 9.46NTGGTIYY110 pKa = 10.51KK111 pKa = 10.51GLLLQNDD118 pKa = 4.35TLNEE122 pKa = 3.69ITRR125 pKa = 11.84LYY127 pKa = 10.8VAFTGEE133 pKa = 3.65QWRR136 pKa = 11.84NSAGGTQILAFSYY149 pKa = 10.46QVGASAITSLTAGAWTSLTSLDD171 pKa = 3.56FSAPVTSGTAGALNGNLPANQRR193 pKa = 11.84RR194 pKa = 11.84RR195 pKa = 11.84TAQFSVAIPAGSYY208 pKa = 10.82IMLRR212 pKa = 11.84WTDD215 pKa = 3.31ADD217 pKa = 3.77EE218 pKa = 4.22QGADD222 pKa = 3.4HH223 pKa = 7.0GLAIDD228 pKa = 4.15DD229 pKa = 4.54LVVSRR234 pKa = 11.84YY235 pKa = 10.0DD236 pKa = 3.59PPQAAADD243 pKa = 4.3AYY245 pKa = 9.07QTSEE249 pKa = 4.28DD250 pKa = 4.03TPLTIAAPGLLANDD264 pKa = 4.41TAHH267 pKa = 6.69QGQPLSAVLVDD278 pKa = 4.18QPSHH282 pKa = 5.87GAVVLNADD290 pKa = 3.74GSFTYY295 pKa = 10.36TPTADD300 pKa = 3.24WNGSDD305 pKa = 4.08SFTYY309 pKa = 8.32TAKK312 pKa = 10.6EE313 pKa = 3.84AGLASAPAAVTLTISPVNDD332 pKa = 3.6APRR335 pKa = 11.84AAADD339 pKa = 3.76GGTTDD344 pKa = 3.59EE345 pKa = 5.28DD346 pKa = 4.15VPLVQPAPGVLDD358 pKa = 3.83NDD360 pKa = 3.61SDD362 pKa = 4.55ADD364 pKa = 3.83GDD366 pKa = 4.31ALTTALVDD374 pKa = 4.44GPSHH378 pKa = 7.31GSLTLNSDD386 pKa = 3.14GSYY389 pKa = 10.06TYY391 pKa = 10.41TPNADD396 pKa = 3.01WNGTDD401 pKa = 3.05NFTYY405 pKa = 9.74TAFDD409 pKa = 3.63GVVNSSVATVTLTVNPAADD428 pKa = 3.95APRR431 pKa = 11.84TTPDD435 pKa = 3.37SYY437 pKa = 11.88DD438 pKa = 3.29VDD440 pKa = 3.9EE441 pKa = 5.54DD442 pKa = 4.05ALLSIAAPGLLEE454 pKa = 4.77NDD456 pKa = 3.27TDD458 pKa = 4.62ADD460 pKa = 4.09GDD462 pKa = 3.87NLTAVWVSDD471 pKa = 3.78PLHH474 pKa = 6.48GVLALNADD482 pKa = 3.68GSYY485 pKa = 10.03TYY487 pKa = 10.79QPDD490 pKa = 3.97ANWNGDD496 pKa = 3.66DD497 pKa = 3.79SFTYY501 pKa = 9.85QASDD505 pKa = 3.45GFLLSAVEE513 pKa = 4.34AVTLHH518 pKa = 5.27VQPINDD524 pKa = 3.97APTAIEE530 pKa = 4.89DD531 pKa = 4.51GYY533 pKa = 9.96STPEE537 pKa = 3.65DD538 pKa = 3.41TQLNVNAPGVLGNDD552 pKa = 3.52TDD554 pKa = 4.51RR555 pKa = 11.84DD556 pKa = 3.86GDD558 pKa = 3.91GLSAHH563 pKa = 6.89LVTDD567 pKa = 4.62ASHH570 pKa = 7.49GDD572 pKa = 3.34LTLNADD578 pKa = 3.86GSFSYY583 pKa = 10.97QPDD586 pKa = 3.79SDD588 pKa = 3.97WNGDD592 pKa = 3.65DD593 pKa = 3.43SFTYY597 pKa = 10.01AAHH600 pKa = 6.92DD601 pKa = 4.05AEE603 pKa = 4.81STSGTVTVEE612 pKa = 3.78LTVNPVNDD620 pKa = 4.44APTATADD627 pKa = 3.5TYY629 pKa = 11.77SLVEE633 pKa = 4.36NGSLSIPANGVLANDD648 pKa = 3.54IDD650 pKa = 3.97VDD652 pKa = 4.25GDD654 pKa = 3.71VLTAVLDD661 pKa = 3.9SSTSHH666 pKa = 7.03GSLTLNTDD674 pKa = 3.09GSFSYY679 pKa = 10.54TPAADD684 pKa = 3.47WNGSDD689 pKa = 4.28SFTYY693 pKa = 10.04HH694 pKa = 6.85ANDD697 pKa = 3.73GAASSEE703 pKa = 4.49TVTVTLTVDD712 pKa = 3.64PDD714 pKa = 3.1NTAPYY719 pKa = 9.87RR720 pKa = 11.84IAPLPDD726 pKa = 3.15QNHH729 pKa = 7.19PARR732 pKa = 11.84TPYY735 pKa = 10.97SFDD738 pKa = 2.84TSAYY742 pKa = 10.35FGDD745 pKa = 4.4SDD747 pKa = 3.92TGDD750 pKa = 3.34VLTFSAQLVDD760 pKa = 5.05GNPLPPWLSCNTASGVLSGTPPLAAIGVYY789 pKa = 9.64SIRR792 pKa = 11.84VTASDD797 pKa = 3.69GSATVSDD804 pKa = 4.95DD805 pKa = 3.48FDD807 pKa = 4.04LTVEE811 pKa = 4.22DD812 pKa = 3.27NPYY815 pKa = 10.75RR816 pKa = 11.84LFIPMVLRR824 pKa = 4.33
Molecular weight: 85.39 kDa
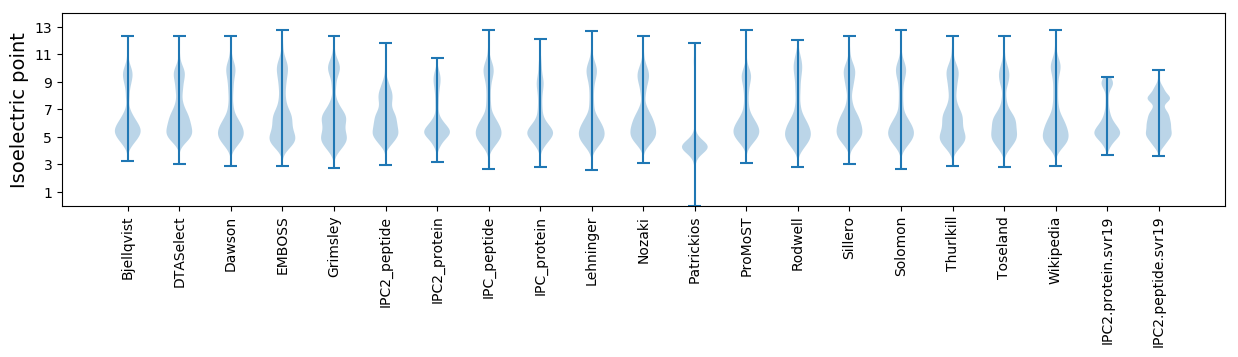
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S7BML4|A0A0S7BML4_9CHLR Transcriptional regulator LacI family OS=Longilinea arvoryzae OX=360412 GN=LARV_02734 PE=4 SV=1
MM1 pKa = 7.42ARR3 pKa = 11.84YY4 pKa = 6.96TGPVCRR10 pKa = 11.84LCRR13 pKa = 11.84RR14 pKa = 11.84EE15 pKa = 3.87NEE17 pKa = 3.97KK18 pKa = 11.08LFLKK22 pKa = 10.73GSRR25 pKa = 11.84CFSAKK30 pKa = 9.99CAFEE34 pKa = 4.07RR35 pKa = 11.84RR36 pKa = 11.84GFAPGQHH43 pKa = 5.91GKK45 pKa = 9.85SGQRR49 pKa = 11.84GGEE52 pKa = 3.91RR53 pKa = 11.84EE54 pKa = 3.86SDD56 pKa = 3.57YY57 pKa = 11.52SRR59 pKa = 11.84QLRR62 pKa = 11.84AKK64 pKa = 8.32QKK66 pKa = 8.27VRR68 pKa = 11.84RR69 pKa = 11.84VYY71 pKa = 10.78GVLEE75 pKa = 3.87RR76 pKa = 11.84QFRR79 pKa = 11.84RR80 pKa = 11.84TFAIARR86 pKa = 11.84KK87 pKa = 7.34TPGLTGLALLVTLEE101 pKa = 3.94MRR103 pKa = 11.84LDD105 pKa = 3.55NVIYY109 pKa = 10.75RR110 pKa = 11.84MGLAEE115 pKa = 4.39NRR117 pKa = 11.84SQARR121 pKa = 11.84QLVNHH126 pKa = 6.07GHH128 pKa = 6.08FLVNGRR134 pKa = 11.84RR135 pKa = 11.84ADD137 pKa = 3.54IPSMILRR144 pKa = 11.84QGDD147 pKa = 4.24VITLHH152 pKa = 5.99EE153 pKa = 4.51SARR156 pKa = 11.84KK157 pKa = 4.98TTYY160 pKa = 9.95FKK162 pKa = 11.03EE163 pKa = 4.12LADD166 pKa = 3.62VAEE169 pKa = 4.42KK170 pKa = 10.35RR171 pKa = 11.84VCASWLDD178 pKa = 3.61RR179 pKa = 11.84DD180 pKa = 4.23LKK182 pKa = 9.85TISGRR187 pKa = 11.84VLRR190 pKa = 11.84SPEE193 pKa = 3.62RR194 pKa = 11.84AEE196 pKa = 3.86IDD198 pKa = 3.6GNLNEE203 pKa = 4.11QLIVEE208 pKa = 4.55YY209 pKa = 10.47YY210 pKa = 10.54SRR212 pKa = 4.63
MM1 pKa = 7.42ARR3 pKa = 11.84YY4 pKa = 6.96TGPVCRR10 pKa = 11.84LCRR13 pKa = 11.84RR14 pKa = 11.84EE15 pKa = 3.87NEE17 pKa = 3.97KK18 pKa = 11.08LFLKK22 pKa = 10.73GSRR25 pKa = 11.84CFSAKK30 pKa = 9.99CAFEE34 pKa = 4.07RR35 pKa = 11.84RR36 pKa = 11.84GFAPGQHH43 pKa = 5.91GKK45 pKa = 9.85SGQRR49 pKa = 11.84GGEE52 pKa = 3.91RR53 pKa = 11.84EE54 pKa = 3.86SDD56 pKa = 3.57YY57 pKa = 11.52SRR59 pKa = 11.84QLRR62 pKa = 11.84AKK64 pKa = 8.32QKK66 pKa = 8.27VRR68 pKa = 11.84RR69 pKa = 11.84VYY71 pKa = 10.78GVLEE75 pKa = 3.87RR76 pKa = 11.84QFRR79 pKa = 11.84RR80 pKa = 11.84TFAIARR86 pKa = 11.84KK87 pKa = 7.34TPGLTGLALLVTLEE101 pKa = 3.94MRR103 pKa = 11.84LDD105 pKa = 3.55NVIYY109 pKa = 10.75RR110 pKa = 11.84MGLAEE115 pKa = 4.39NRR117 pKa = 11.84SQARR121 pKa = 11.84QLVNHH126 pKa = 6.07GHH128 pKa = 6.08FLVNGRR134 pKa = 11.84RR135 pKa = 11.84ADD137 pKa = 3.54IPSMILRR144 pKa = 11.84QGDD147 pKa = 4.24VITLHH152 pKa = 5.99EE153 pKa = 4.51SARR156 pKa = 11.84KK157 pKa = 4.98TTYY160 pKa = 9.95FKK162 pKa = 11.03EE163 pKa = 4.12LADD166 pKa = 3.62VAEE169 pKa = 4.42KK170 pKa = 10.35RR171 pKa = 11.84VCASWLDD178 pKa = 3.61RR179 pKa = 11.84DD180 pKa = 4.23LKK182 pKa = 9.85TISGRR187 pKa = 11.84VLRR190 pKa = 11.84SPEE193 pKa = 3.62RR194 pKa = 11.84AEE196 pKa = 3.86IDD198 pKa = 3.6GNLNEE203 pKa = 4.11QLIVEE208 pKa = 4.55YY209 pKa = 10.47YY210 pKa = 10.54SRR212 pKa = 4.63
Molecular weight: 24.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1319558 |
27 |
2980 |
340.9 |
37.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.897 ± 0.053 | 0.903 ± 0.015 |
5.201 ± 0.027 | 5.804 ± 0.04 |
3.968 ± 0.03 | 7.785 ± 0.035 |
1.911 ± 0.017 | 5.966 ± 0.039 |
3.465 ± 0.036 | 11.106 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.362 ± 0.019 | 3.41 ± 0.028 |
5.255 ± 0.034 | 4.037 ± 0.025 |
5.975 ± 0.037 | 5.828 ± 0.038 |
5.504 ± 0.05 | 7.101 ± 0.031 |
1.569 ± 0.017 | 2.954 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
