
Halorussus sp. MSC15.2
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halobacteriaceae; Halorussus; unclassified Halorussus
Average proteome isoelectric point is 4.97
Get precalculated fractions of proteins
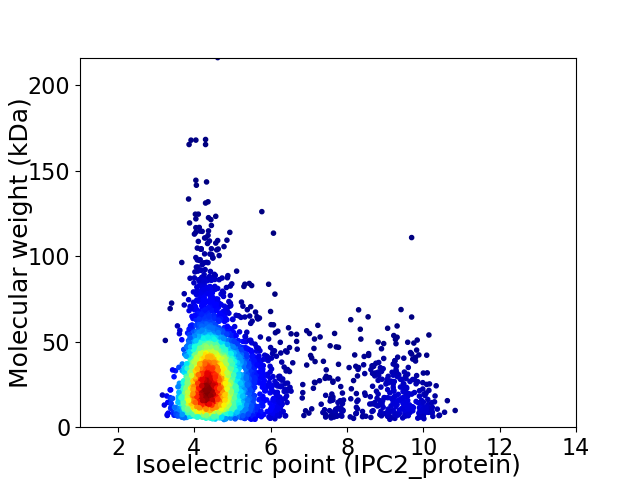
Virtual 2D-PAGE plot for 4072 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G3ZHY4|A0A6G3ZHY4_9EURY Metal-dependent hydrolase OS=Halorussus sp. MSC15.2 OX=2283638 GN=FXF75_04805 PE=4 SV=1
MM1 pKa = 7.29NRR3 pKa = 11.84RR4 pKa = 11.84TFLSALATAAGSSAAGIEE22 pKa = 3.94LTGRR26 pKa = 11.84SRR28 pKa = 11.84AASGRR33 pKa = 11.84IPEE36 pKa = 4.44LEE38 pKa = 4.42AYY40 pKa = 7.58STSSLVTYY48 pKa = 10.06DD49 pKa = 3.91YY50 pKa = 11.84APLTDD55 pKa = 4.45DD56 pKa = 3.26AHH58 pKa = 5.63VAVWAEE64 pKa = 4.05DD65 pKa = 3.42TATNGDD71 pKa = 3.81GDD73 pKa = 3.99TSADD77 pKa = 3.21AVSYY81 pKa = 11.54GEE83 pKa = 4.3DD84 pKa = 3.33TPIPVVASDD93 pKa = 3.48WNVVGFGSMLVTDD106 pKa = 5.81ADD108 pKa = 4.36TDD110 pKa = 3.6WQTGNEE116 pKa = 3.97EE117 pKa = 3.91FVLNVWDD124 pKa = 4.58DD125 pKa = 3.73ALGGSGTVLWDD136 pKa = 3.6EE137 pKa = 4.43GHH139 pKa = 5.63GQYY142 pKa = 8.64YY143 pKa = 8.68TLSKK147 pKa = 10.28FAEE150 pKa = 3.98FDD152 pKa = 3.63GYY154 pKa = 11.71AEE156 pKa = 4.17NNGYY160 pKa = 9.99AVSATTSLTADD171 pKa = 4.35LSAADD176 pKa = 3.95AAVITSPSTAFTQSEE191 pKa = 4.4LSALGDD197 pKa = 3.78FVSNGGWLFLHH208 pKa = 6.8DD209 pKa = 4.62QSDD212 pKa = 3.46HH213 pKa = 6.44SNYY216 pKa = 10.84DD217 pKa = 3.02EE218 pKa = 4.15TANLNDD224 pKa = 3.67VASHH228 pKa = 6.81LEE230 pKa = 3.67LAFRR234 pKa = 11.84FNDD237 pKa = 3.57DD238 pKa = 3.07QVTDD242 pKa = 4.53EE243 pKa = 4.54IRR245 pKa = 11.84NGGEE249 pKa = 3.62PYY251 pKa = 10.59QPVTTQFNTAFSYY264 pKa = 10.84FGDD267 pKa = 3.66RR268 pKa = 11.84DD269 pKa = 3.94GLGLDD274 pKa = 3.52PEE276 pKa = 4.38KK277 pKa = 10.3TYY279 pKa = 10.14TVSVTEE285 pKa = 4.71VIDD288 pKa = 4.0GDD290 pKa = 4.36TVKK293 pKa = 9.43VTFDD297 pKa = 4.15DD298 pKa = 4.22GTTEE302 pKa = 4.1NIRR305 pKa = 11.84ILGTDD310 pKa = 3.48TPEE313 pKa = 4.09KK314 pKa = 10.5SSNSQYY320 pKa = 10.93EE321 pKa = 4.15RR322 pKa = 11.84VEE324 pKa = 3.76EE325 pKa = 4.26WEE327 pKa = 4.74GIEE330 pKa = 4.49SNTYY334 pKa = 10.44LEE336 pKa = 4.76ARR338 pKa = 11.84ADD340 pKa = 3.58DD341 pKa = 3.57ATNFGKK347 pKa = 10.25SEE349 pKa = 4.21LGGKK353 pKa = 9.45SVDD356 pKa = 4.37LAFDD360 pKa = 4.1DD361 pKa = 4.4NEE363 pKa = 4.19PVRR366 pKa = 11.84DD367 pKa = 3.56AFDD370 pKa = 3.51RR371 pKa = 11.84VLGYY375 pKa = 9.93IYY377 pKa = 10.82YY378 pKa = 10.22DD379 pKa = 3.29ASGDD383 pKa = 3.76GNRR386 pKa = 11.84DD387 pKa = 2.74ANYY390 pKa = 10.3NHH392 pKa = 7.14RR393 pKa = 11.84LVKK396 pKa = 10.44NGHH399 pKa = 5.69ARR401 pKa = 11.84VYY403 pKa = 10.42DD404 pKa = 3.68SSFAKK409 pKa = 10.27HH410 pKa = 6.32DD411 pKa = 3.79AFVDD415 pKa = 3.76SEE417 pKa = 4.49TTARR421 pKa = 11.84ADD423 pKa = 3.62GTQVWAEE430 pKa = 4.07SDD432 pKa = 3.51PGNSTEE438 pKa = 4.83IRR440 pKa = 11.84DD441 pKa = 3.97NPVDD445 pKa = 4.9DD446 pKa = 5.61LFFPQTASVRR456 pKa = 11.84TASGGVPDD464 pKa = 4.24SRR466 pKa = 11.84VPVYY470 pKa = 10.84AEE472 pKa = 3.84STATQDD478 pKa = 3.51LDD480 pKa = 3.29GGYY483 pKa = 9.86AYY485 pKa = 10.83SGDD488 pKa = 3.83IPLVAVDD495 pKa = 3.71EE496 pKa = 4.58DD497 pKa = 4.63ANVGVVGGPFIDD509 pKa = 3.81EE510 pKa = 4.45SYY512 pKa = 10.37EE513 pKa = 3.96KK514 pKa = 10.95SEE516 pKa = 4.71GYY518 pKa = 8.14STDD521 pKa = 2.89TSSYY525 pKa = 11.07GNFAFLTNLVDD536 pKa = 3.97YY537 pKa = 10.85LADD540 pKa = 3.44TSGDD544 pKa = 3.62VLVDD548 pKa = 3.81GGHH551 pKa = 5.71GQFGASYY558 pKa = 9.97GLSAEE563 pKa = 3.9DD564 pKa = 2.77TAYY567 pKa = 10.94YY568 pKa = 9.5MRR570 pKa = 11.84YY571 pKa = 10.21LEE573 pKa = 4.58GQDD576 pKa = 3.38VALEE580 pKa = 4.2GVNEE584 pKa = 3.97FTGANLDD591 pKa = 3.44GARR594 pKa = 11.84AVVVTTPPEE603 pKa = 3.93SFTQSEE609 pKa = 3.92IDD611 pKa = 3.41ALNDD615 pKa = 3.48FVADD619 pKa = 4.2GGAVVLVGSGKK630 pKa = 6.84TTAAARR636 pKa = 11.84ANVNDD641 pKa = 4.25LADD644 pKa = 4.06GLGTDD649 pKa = 4.16LQVNADD655 pKa = 3.93RR656 pKa = 11.84VLDD659 pKa = 3.91DD660 pKa = 4.02TNSVATSPEE669 pKa = 4.1VPEE672 pKa = 4.08TTVFDD677 pKa = 4.02GSFPLFDD684 pKa = 5.11AYY686 pKa = 9.97TPGTSSDD693 pKa = 3.7YY694 pKa = 10.94SVSIPTISEE703 pKa = 4.32DD704 pKa = 4.15GSTLDD709 pKa = 5.2DD710 pKa = 4.27EE711 pKa = 4.9YY712 pKa = 11.99VDD714 pKa = 3.84VKK716 pKa = 11.39NDD718 pKa = 3.15GSSAQDD724 pKa = 3.27LTGWTLEE731 pKa = 4.2DD732 pKa = 3.93EE733 pKa = 4.98AGHH736 pKa = 6.36TYY738 pKa = 10.47GFPDD742 pKa = 4.54GFSLGAGDD750 pKa = 3.9TVRR753 pKa = 11.84VHH755 pKa = 6.41TGSGTDD761 pKa = 3.41SSTDD765 pKa = 3.67LYY767 pKa = 9.93WDD769 pKa = 3.62RR770 pKa = 11.84GSAVWNDD777 pKa = 3.58GGDD780 pKa = 3.51TASVYY785 pKa = 11.35DD786 pKa = 4.24DD787 pKa = 4.53AGNLAAEE794 pKa = 4.39KK795 pKa = 9.63TYY797 pKa = 8.56PTNDD801 pKa = 3.52SDD803 pKa = 4.88SKK805 pKa = 10.81IAVKK809 pKa = 9.33TVSEE813 pKa = 4.43AGSTLNDD820 pKa = 3.24EE821 pKa = 4.64YY822 pKa = 11.84VDD824 pKa = 4.22FEE826 pKa = 4.56NTGSSAQDD834 pKa = 3.23LTGWTVEE841 pKa = 4.33DD842 pKa = 4.61EE843 pKa = 4.36ADD845 pKa = 3.34HH846 pKa = 6.79TYY848 pKa = 11.14GFPDD852 pKa = 4.39GFSLGAGEE860 pKa = 4.24TVRR863 pKa = 11.84LHH865 pKa = 6.76SGDD868 pKa = 3.78GTDD871 pKa = 4.94SATDD875 pKa = 4.24LYY877 pKa = 10.73WGGSYY882 pKa = 9.51VWNDD886 pKa = 3.08GGDD889 pKa = 3.64TVSLYY894 pKa = 10.84DD895 pKa = 4.56AGGTLHH901 pKa = 6.62TEE903 pKa = 3.69YY904 pKa = 10.85SYY906 pKa = 12.1
MM1 pKa = 7.29NRR3 pKa = 11.84RR4 pKa = 11.84TFLSALATAAGSSAAGIEE22 pKa = 3.94LTGRR26 pKa = 11.84SRR28 pKa = 11.84AASGRR33 pKa = 11.84IPEE36 pKa = 4.44LEE38 pKa = 4.42AYY40 pKa = 7.58STSSLVTYY48 pKa = 10.06DD49 pKa = 3.91YY50 pKa = 11.84APLTDD55 pKa = 4.45DD56 pKa = 3.26AHH58 pKa = 5.63VAVWAEE64 pKa = 4.05DD65 pKa = 3.42TATNGDD71 pKa = 3.81GDD73 pKa = 3.99TSADD77 pKa = 3.21AVSYY81 pKa = 11.54GEE83 pKa = 4.3DD84 pKa = 3.33TPIPVVASDD93 pKa = 3.48WNVVGFGSMLVTDD106 pKa = 5.81ADD108 pKa = 4.36TDD110 pKa = 3.6WQTGNEE116 pKa = 3.97EE117 pKa = 3.91FVLNVWDD124 pKa = 4.58DD125 pKa = 3.73ALGGSGTVLWDD136 pKa = 3.6EE137 pKa = 4.43GHH139 pKa = 5.63GQYY142 pKa = 8.64YY143 pKa = 8.68TLSKK147 pKa = 10.28FAEE150 pKa = 3.98FDD152 pKa = 3.63GYY154 pKa = 11.71AEE156 pKa = 4.17NNGYY160 pKa = 9.99AVSATTSLTADD171 pKa = 4.35LSAADD176 pKa = 3.95AAVITSPSTAFTQSEE191 pKa = 4.4LSALGDD197 pKa = 3.78FVSNGGWLFLHH208 pKa = 6.8DD209 pKa = 4.62QSDD212 pKa = 3.46HH213 pKa = 6.44SNYY216 pKa = 10.84DD217 pKa = 3.02EE218 pKa = 4.15TANLNDD224 pKa = 3.67VASHH228 pKa = 6.81LEE230 pKa = 3.67LAFRR234 pKa = 11.84FNDD237 pKa = 3.57DD238 pKa = 3.07QVTDD242 pKa = 4.53EE243 pKa = 4.54IRR245 pKa = 11.84NGGEE249 pKa = 3.62PYY251 pKa = 10.59QPVTTQFNTAFSYY264 pKa = 10.84FGDD267 pKa = 3.66RR268 pKa = 11.84DD269 pKa = 3.94GLGLDD274 pKa = 3.52PEE276 pKa = 4.38KK277 pKa = 10.3TYY279 pKa = 10.14TVSVTEE285 pKa = 4.71VIDD288 pKa = 4.0GDD290 pKa = 4.36TVKK293 pKa = 9.43VTFDD297 pKa = 4.15DD298 pKa = 4.22GTTEE302 pKa = 4.1NIRR305 pKa = 11.84ILGTDD310 pKa = 3.48TPEE313 pKa = 4.09KK314 pKa = 10.5SSNSQYY320 pKa = 10.93EE321 pKa = 4.15RR322 pKa = 11.84VEE324 pKa = 3.76EE325 pKa = 4.26WEE327 pKa = 4.74GIEE330 pKa = 4.49SNTYY334 pKa = 10.44LEE336 pKa = 4.76ARR338 pKa = 11.84ADD340 pKa = 3.58DD341 pKa = 3.57ATNFGKK347 pKa = 10.25SEE349 pKa = 4.21LGGKK353 pKa = 9.45SVDD356 pKa = 4.37LAFDD360 pKa = 4.1DD361 pKa = 4.4NEE363 pKa = 4.19PVRR366 pKa = 11.84DD367 pKa = 3.56AFDD370 pKa = 3.51RR371 pKa = 11.84VLGYY375 pKa = 9.93IYY377 pKa = 10.82YY378 pKa = 10.22DD379 pKa = 3.29ASGDD383 pKa = 3.76GNRR386 pKa = 11.84DD387 pKa = 2.74ANYY390 pKa = 10.3NHH392 pKa = 7.14RR393 pKa = 11.84LVKK396 pKa = 10.44NGHH399 pKa = 5.69ARR401 pKa = 11.84VYY403 pKa = 10.42DD404 pKa = 3.68SSFAKK409 pKa = 10.27HH410 pKa = 6.32DD411 pKa = 3.79AFVDD415 pKa = 3.76SEE417 pKa = 4.49TTARR421 pKa = 11.84ADD423 pKa = 3.62GTQVWAEE430 pKa = 4.07SDD432 pKa = 3.51PGNSTEE438 pKa = 4.83IRR440 pKa = 11.84DD441 pKa = 3.97NPVDD445 pKa = 4.9DD446 pKa = 5.61LFFPQTASVRR456 pKa = 11.84TASGGVPDD464 pKa = 4.24SRR466 pKa = 11.84VPVYY470 pKa = 10.84AEE472 pKa = 3.84STATQDD478 pKa = 3.51LDD480 pKa = 3.29GGYY483 pKa = 9.86AYY485 pKa = 10.83SGDD488 pKa = 3.83IPLVAVDD495 pKa = 3.71EE496 pKa = 4.58DD497 pKa = 4.63ANVGVVGGPFIDD509 pKa = 3.81EE510 pKa = 4.45SYY512 pKa = 10.37EE513 pKa = 3.96KK514 pKa = 10.95SEE516 pKa = 4.71GYY518 pKa = 8.14STDD521 pKa = 2.89TSSYY525 pKa = 11.07GNFAFLTNLVDD536 pKa = 3.97YY537 pKa = 10.85LADD540 pKa = 3.44TSGDD544 pKa = 3.62VLVDD548 pKa = 3.81GGHH551 pKa = 5.71GQFGASYY558 pKa = 9.97GLSAEE563 pKa = 3.9DD564 pKa = 2.77TAYY567 pKa = 10.94YY568 pKa = 9.5MRR570 pKa = 11.84YY571 pKa = 10.21LEE573 pKa = 4.58GQDD576 pKa = 3.38VALEE580 pKa = 4.2GVNEE584 pKa = 3.97FTGANLDD591 pKa = 3.44GARR594 pKa = 11.84AVVVTTPPEE603 pKa = 3.93SFTQSEE609 pKa = 3.92IDD611 pKa = 3.41ALNDD615 pKa = 3.48FVADD619 pKa = 4.2GGAVVLVGSGKK630 pKa = 6.84TTAAARR636 pKa = 11.84ANVNDD641 pKa = 4.25LADD644 pKa = 4.06GLGTDD649 pKa = 4.16LQVNADD655 pKa = 3.93RR656 pKa = 11.84VLDD659 pKa = 3.91DD660 pKa = 4.02TNSVATSPEE669 pKa = 4.1VPEE672 pKa = 4.08TTVFDD677 pKa = 4.02GSFPLFDD684 pKa = 5.11AYY686 pKa = 9.97TPGTSSDD693 pKa = 3.7YY694 pKa = 10.94SVSIPTISEE703 pKa = 4.32DD704 pKa = 4.15GSTLDD709 pKa = 5.2DD710 pKa = 4.27EE711 pKa = 4.9YY712 pKa = 11.99VDD714 pKa = 3.84VKK716 pKa = 11.39NDD718 pKa = 3.15GSSAQDD724 pKa = 3.27LTGWTLEE731 pKa = 4.2DD732 pKa = 3.93EE733 pKa = 4.98AGHH736 pKa = 6.36TYY738 pKa = 10.47GFPDD742 pKa = 4.54GFSLGAGDD750 pKa = 3.9TVRR753 pKa = 11.84VHH755 pKa = 6.41TGSGTDD761 pKa = 3.41SSTDD765 pKa = 3.67LYY767 pKa = 9.93WDD769 pKa = 3.62RR770 pKa = 11.84GSAVWNDD777 pKa = 3.58GGDD780 pKa = 3.51TASVYY785 pKa = 11.35DD786 pKa = 4.24DD787 pKa = 4.53AGNLAAEE794 pKa = 4.39KK795 pKa = 9.63TYY797 pKa = 8.56PTNDD801 pKa = 3.52SDD803 pKa = 4.88SKK805 pKa = 10.81IAVKK809 pKa = 9.33TVSEE813 pKa = 4.43AGSTLNDD820 pKa = 3.24EE821 pKa = 4.64YY822 pKa = 11.84VDD824 pKa = 4.22FEE826 pKa = 4.56NTGSSAQDD834 pKa = 3.23LTGWTVEE841 pKa = 4.33DD842 pKa = 4.61EE843 pKa = 4.36ADD845 pKa = 3.34HH846 pKa = 6.79TYY848 pKa = 11.14GFPDD852 pKa = 4.39GFSLGAGEE860 pKa = 4.24TVRR863 pKa = 11.84LHH865 pKa = 6.76SGDD868 pKa = 3.78GTDD871 pKa = 4.94SATDD875 pKa = 4.24LYY877 pKa = 10.73WGGSYY882 pKa = 9.51VWNDD886 pKa = 3.08GGDD889 pKa = 3.64TVSLYY894 pKa = 10.84DD895 pKa = 4.56AGGTLHH901 pKa = 6.62TEE903 pKa = 3.69YY904 pKa = 10.85SYY906 pKa = 12.1
Molecular weight: 96.36 kDa
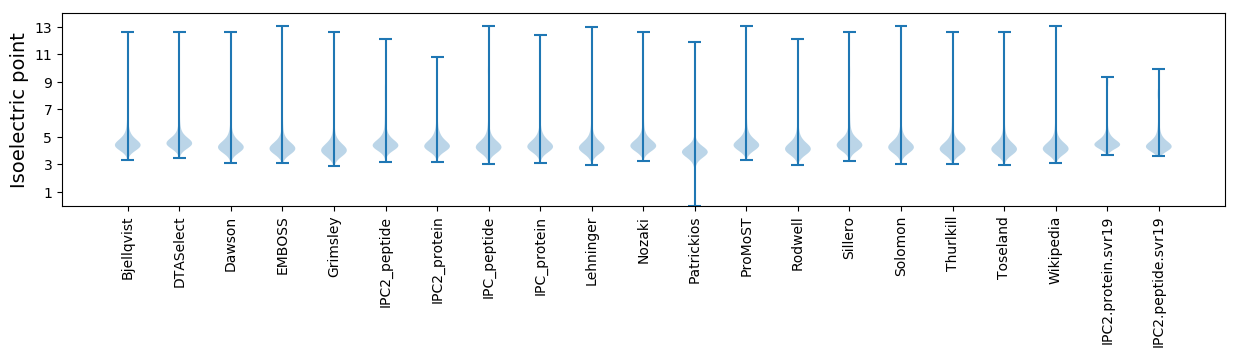
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G3ZER7|A0A6G3ZER7_9EURY Uncharacterized protein OS=Halorussus sp. MSC15.2 OX=2283638 GN=FXF75_00940 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 10.46LNTRR6 pKa = 11.84IRR8 pKa = 11.84RR9 pKa = 11.84TGPASRR15 pKa = 11.84TRR17 pKa = 11.84RR18 pKa = 11.84GRR20 pKa = 11.84IASGVTRR27 pKa = 11.84FALPVATLLALVASALLTGVTFATVAALLPMPVPLLLGGWVAAIGLTFALPLVVVRR83 pKa = 11.84GVVALLEE90 pKa = 4.35RR91 pKa = 11.84FQQ93 pKa = 4.59
MM1 pKa = 7.63KK2 pKa = 10.46LNTRR6 pKa = 11.84IRR8 pKa = 11.84RR9 pKa = 11.84TGPASRR15 pKa = 11.84TRR17 pKa = 11.84RR18 pKa = 11.84GRR20 pKa = 11.84IASGVTRR27 pKa = 11.84FALPVATLLALVASALLTGVTFATVAALLPMPVPLLLGGWVAAIGLTFALPLVVVRR83 pKa = 11.84GVVALLEE90 pKa = 4.35RR91 pKa = 11.84FQQ93 pKa = 4.59
Molecular weight: 9.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1157722 |
44 |
2031 |
284.3 |
30.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.692 ± 0.054 | 0.662 ± 0.013 |
8.269 ± 0.044 | 8.683 ± 0.06 |
3.426 ± 0.027 | 8.554 ± 0.041 |
1.895 ± 0.021 | 3.383 ± 0.029 |
2.085 ± 0.026 | 8.677 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.709 ± 0.016 | 2.59 ± 0.024 |
4.527 ± 0.025 | 2.496 ± 0.024 |
6.71 ± 0.039 | 5.847 ± 0.041 |
6.537 ± 0.049 | 9.306 ± 0.042 |
1.186 ± 0.013 | 2.769 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
