
Caenorhabditis brenneri (Nematode worm)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Rhabditina; Rhabditomorpha; Rhabditoidea; Rhabditidae; Peloderinae; Caenorhabditis
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
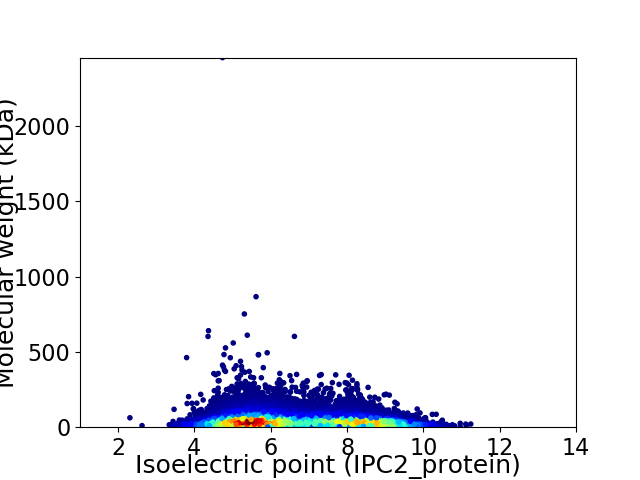
Virtual 2D-PAGE plot for 29982 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G0P6K2|G0P6K2_CAEBE CBN-TIN-10 protein OS=Caenorhabditis brenneri OX=135651 GN=Cbn-tin-10 PE=4 SV=1
MM1 pKa = 7.7KK2 pKa = 10.14FQLVFLLAALIGSSQAQGLPGVICGITGILCPPTTVAPTTTTTTVPTTTTASDD55 pKa = 3.19TSEE58 pKa = 3.66ICFTIPGNNVNIVIDD73 pKa = 4.05PTQLTLTEE81 pKa = 4.26IGQIKK86 pKa = 9.71QLVEE90 pKa = 4.23DD91 pKa = 4.04SLANILGSLGGTINGALNGLVGGLTGILNPATTPTPTGDD130 pKa = 3.33GQICVVIPNSDD141 pKa = 3.16IDD143 pKa = 3.7EE144 pKa = 4.66LEE146 pKa = 4.21AFLEE150 pKa = 4.6DD151 pKa = 4.02VFSLLTAFFNCLKK164 pKa = 10.66DD165 pKa = 4.13LEE167 pKa = 4.45EE168 pKa = 4.22TTAQFQAEE176 pKa = 4.27VDD178 pKa = 3.87RR179 pKa = 11.84VRR181 pKa = 11.84MEE183 pKa = 3.62IDD185 pKa = 3.07TLVARR190 pKa = 11.84FNNCDD195 pKa = 2.92IVTCYY200 pKa = 10.69LGVANDD206 pKa = 3.45AYY208 pKa = 10.38EE209 pKa = 4.23LVRR212 pKa = 11.84SLPARR217 pKa = 11.84FLTYY221 pKa = 10.59PNTVRR226 pKa = 11.84DD227 pKa = 4.34NINQCVTEE235 pKa = 4.54TINAA239 pKa = 3.89
MM1 pKa = 7.7KK2 pKa = 10.14FQLVFLLAALIGSSQAQGLPGVICGITGILCPPTTVAPTTTTTTVPTTTTASDD55 pKa = 3.19TSEE58 pKa = 3.66ICFTIPGNNVNIVIDD73 pKa = 4.05PTQLTLTEE81 pKa = 4.26IGQIKK86 pKa = 9.71QLVEE90 pKa = 4.23DD91 pKa = 4.04SLANILGSLGGTINGALNGLVGGLTGILNPATTPTPTGDD130 pKa = 3.33GQICVVIPNSDD141 pKa = 3.16IDD143 pKa = 3.7EE144 pKa = 4.66LEE146 pKa = 4.21AFLEE150 pKa = 4.6DD151 pKa = 4.02VFSLLTAFFNCLKK164 pKa = 10.66DD165 pKa = 4.13LEE167 pKa = 4.45EE168 pKa = 4.22TTAQFQAEE176 pKa = 4.27VDD178 pKa = 3.87RR179 pKa = 11.84VRR181 pKa = 11.84MEE183 pKa = 3.62IDD185 pKa = 3.07TLVARR190 pKa = 11.84FNNCDD195 pKa = 2.92IVTCYY200 pKa = 10.69LGVANDD206 pKa = 3.45AYY208 pKa = 10.38EE209 pKa = 4.23LVRR212 pKa = 11.84SLPARR217 pKa = 11.84FLTYY221 pKa = 10.59PNTVRR226 pKa = 11.84DD227 pKa = 4.34NINQCVTEE235 pKa = 4.54TINAA239 pKa = 3.89
Molecular weight: 25.36 kDa
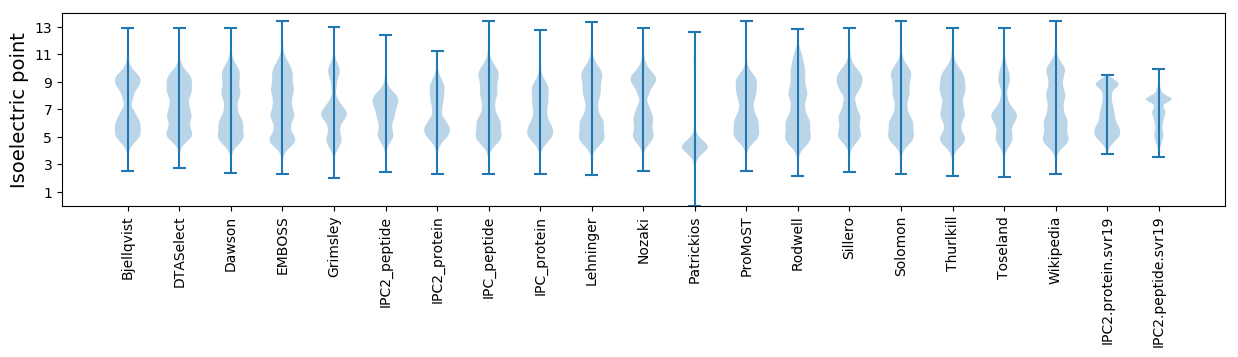
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G0PHB7|G0PHB7_CAEBE Uncharacterized protein OS=Caenorhabditis brenneri OX=135651 GN=CAEBREN_02941 PE=4 SV=1
MM1 pKa = 7.56KK2 pKa = 10.44SNRR5 pKa = 11.84SNSRR9 pKa = 11.84GRR11 pKa = 11.84QRR13 pKa = 11.84DD14 pKa = 3.38SKK16 pKa = 11.55GRR18 pKa = 11.84FTPKK22 pKa = 9.65RR23 pKa = 11.84KK24 pKa = 7.58QTIVEE29 pKa = 4.4RR30 pKa = 11.84SRR32 pKa = 11.84SRR34 pKa = 11.84SRR36 pKa = 11.84SSSRR40 pKa = 11.84GGHH43 pKa = 4.68RR44 pKa = 11.84RR45 pKa = 11.84GRR47 pKa = 11.84SSTRR51 pKa = 11.84KK52 pKa = 8.5SLAVSLHH59 pKa = 5.07TARR62 pKa = 11.84SPSVSSQRR70 pKa = 11.84SSSRR74 pKa = 11.84RR75 pKa = 11.84SVMRR79 pKa = 11.84SSRR82 pKa = 11.84STSSRR87 pKa = 11.84RR88 pKa = 11.84SSSRR92 pKa = 11.84GSVYY96 pKa = 10.76GKK98 pKa = 8.4TGRR101 pKa = 11.84SRR103 pKa = 11.84SSHH106 pKa = 5.67RR107 pKa = 11.84SSRR110 pKa = 11.84SRR112 pKa = 11.84SASRR116 pKa = 11.84SATRR120 pKa = 11.84SSSSRR125 pKa = 11.84SRR127 pKa = 11.84SSSRR131 pKa = 11.84SRR133 pKa = 11.84RR134 pKa = 11.84SASQKK139 pKa = 8.84KK140 pKa = 9.4KK141 pKa = 9.3KK142 pKa = 9.12VVRR145 pKa = 11.84RR146 pKa = 11.84GRR148 pKa = 11.84AARR151 pKa = 11.84SAKK154 pKa = 8.3STKK157 pKa = 8.9SLKK160 pKa = 9.03TSRR163 pKa = 11.84SRR165 pKa = 11.84SRR167 pKa = 11.84SQGPSEE173 pKa = 4.28KK174 pKa = 10.12KK175 pKa = 10.14SRR177 pKa = 11.84TPVARR182 pKa = 11.84STRR185 pKa = 11.84RR186 pKa = 11.84RR187 pKa = 11.84AA188 pKa = 3.07
MM1 pKa = 7.56KK2 pKa = 10.44SNRR5 pKa = 11.84SNSRR9 pKa = 11.84GRR11 pKa = 11.84QRR13 pKa = 11.84DD14 pKa = 3.38SKK16 pKa = 11.55GRR18 pKa = 11.84FTPKK22 pKa = 9.65RR23 pKa = 11.84KK24 pKa = 7.58QTIVEE29 pKa = 4.4RR30 pKa = 11.84SRR32 pKa = 11.84SRR34 pKa = 11.84SRR36 pKa = 11.84SSSRR40 pKa = 11.84GGHH43 pKa = 4.68RR44 pKa = 11.84RR45 pKa = 11.84GRR47 pKa = 11.84SSTRR51 pKa = 11.84KK52 pKa = 8.5SLAVSLHH59 pKa = 5.07TARR62 pKa = 11.84SPSVSSQRR70 pKa = 11.84SSSRR74 pKa = 11.84RR75 pKa = 11.84SVMRR79 pKa = 11.84SSRR82 pKa = 11.84STSSRR87 pKa = 11.84RR88 pKa = 11.84SSSRR92 pKa = 11.84GSVYY96 pKa = 10.76GKK98 pKa = 8.4TGRR101 pKa = 11.84SRR103 pKa = 11.84SSHH106 pKa = 5.67RR107 pKa = 11.84SSRR110 pKa = 11.84SRR112 pKa = 11.84SASRR116 pKa = 11.84SATRR120 pKa = 11.84SSSSRR125 pKa = 11.84SRR127 pKa = 11.84SSSRR131 pKa = 11.84SRR133 pKa = 11.84RR134 pKa = 11.84SASQKK139 pKa = 8.84KK140 pKa = 9.4KK141 pKa = 9.3KK142 pKa = 9.12VVRR145 pKa = 11.84RR146 pKa = 11.84GRR148 pKa = 11.84AARR151 pKa = 11.84SAKK154 pKa = 8.3STKK157 pKa = 8.9SLKK160 pKa = 9.03TSRR163 pKa = 11.84SRR165 pKa = 11.84SRR167 pKa = 11.84SQGPSEE173 pKa = 4.28KK174 pKa = 10.12KK175 pKa = 10.14SRR177 pKa = 11.84TPVARR182 pKa = 11.84STRR185 pKa = 11.84RR186 pKa = 11.84RR187 pKa = 11.84AA188 pKa = 3.07
Molecular weight: 21.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11989625 |
8 |
22383 |
399.9 |
45.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.231 ± 0.018 | 1.926 ± 0.012 |
5.433 ± 0.014 | 7.157 ± 0.024 |
4.568 ± 0.014 | 5.297 ± 0.018 |
2.303 ± 0.009 | 5.864 ± 0.012 |
6.602 ± 0.019 | 8.483 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.638 ± 0.007 | 4.843 ± 0.01 |
5.131 ± 0.022 | 4.166 ± 0.015 |
5.34 ± 0.014 | 7.775 ± 0.021 |
5.817 ± 0.018 | 6.185 ± 0.012 |
1.109 ± 0.005 | 3.098 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
