
Thermobifida phage P1312
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
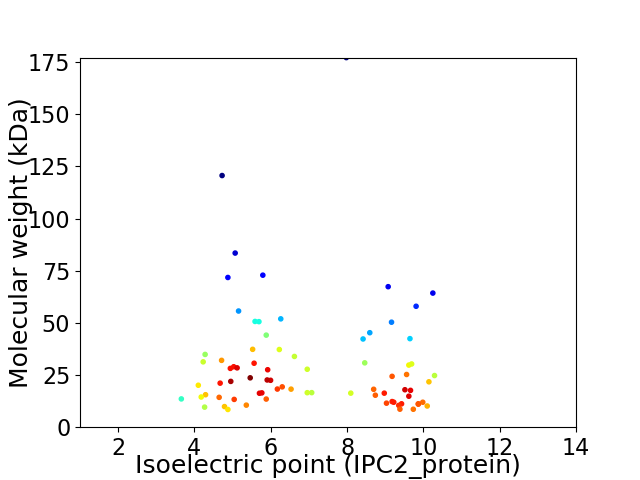
Virtual 2D-PAGE plot for 77 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0R8V9X2|A0A0R8V9X2_9CAUD Transposase OS=Thermobifida phage P1312 OX=1661715 GN=P1312_049 PE=3 SV=1
MM1 pKa = 7.95EE2 pKa = 6.29DD3 pKa = 4.22LMHH6 pKa = 7.29PPYY9 pKa = 10.08QQPPHH14 pKa = 6.8SSPVRR19 pKa = 11.84QQGMSAGAKK28 pKa = 9.24VALAGCGGCGALIGLVLMAGCVAAIVAPADD58 pKa = 3.67PAPPAAEE65 pKa = 4.13SSASGSASPEE75 pKa = 3.92EE76 pKa = 4.47SVSPSPEE83 pKa = 3.73PSEE86 pKa = 4.16TPEE89 pKa = 4.52DD90 pKa = 3.59EE91 pKa = 4.65VSFEE95 pKa = 4.18EE96 pKa = 5.51LGFPPALEE104 pKa = 4.37GEE106 pKa = 4.16EE107 pKa = 3.9RR108 pKa = 11.84AAYY111 pKa = 9.76LADD114 pKa = 3.76VRR116 pKa = 11.84DD117 pKa = 3.81LDD119 pKa = 4.01RR120 pKa = 11.84GFVFPDD126 pKa = 3.21EE127 pKa = 4.89DD128 pKa = 4.04SVVARR133 pKa = 11.84GQDD136 pKa = 3.33VCVTLRR142 pKa = 11.84DD143 pKa = 3.74EE144 pKa = 4.97GEE146 pKa = 3.98QAAIDD151 pKa = 4.14YY152 pKa = 10.84IEE154 pKa = 5.64AIWLHH159 pKa = 6.11EE160 pKa = 4.91DD161 pKa = 3.27YY162 pKa = 11.4DD163 pKa = 4.06FGRR166 pKa = 11.84RR167 pKa = 11.84PTTRR171 pKa = 11.84EE172 pKa = 3.46EE173 pKa = 4.06AEE175 pKa = 3.89QLLGIVHH182 pKa = 6.36EE183 pKa = 4.95HH184 pKa = 6.18VCPDD188 pKa = 2.91WW189 pKa = 5.83
MM1 pKa = 7.95EE2 pKa = 6.29DD3 pKa = 4.22LMHH6 pKa = 7.29PPYY9 pKa = 10.08QQPPHH14 pKa = 6.8SSPVRR19 pKa = 11.84QQGMSAGAKK28 pKa = 9.24VALAGCGGCGALIGLVLMAGCVAAIVAPADD58 pKa = 3.67PAPPAAEE65 pKa = 4.13SSASGSASPEE75 pKa = 3.92EE76 pKa = 4.47SVSPSPEE83 pKa = 3.73PSEE86 pKa = 4.16TPEE89 pKa = 4.52DD90 pKa = 3.59EE91 pKa = 4.65VSFEE95 pKa = 4.18EE96 pKa = 5.51LGFPPALEE104 pKa = 4.37GEE106 pKa = 4.16EE107 pKa = 3.9RR108 pKa = 11.84AAYY111 pKa = 9.76LADD114 pKa = 3.76VRR116 pKa = 11.84DD117 pKa = 3.81LDD119 pKa = 4.01RR120 pKa = 11.84GFVFPDD126 pKa = 3.21EE127 pKa = 4.89DD128 pKa = 4.04SVVARR133 pKa = 11.84GQDD136 pKa = 3.33VCVTLRR142 pKa = 11.84DD143 pKa = 3.74EE144 pKa = 4.97GEE146 pKa = 3.98QAAIDD151 pKa = 4.14YY152 pKa = 10.84IEE154 pKa = 5.64AIWLHH159 pKa = 6.11EE160 pKa = 4.91DD161 pKa = 3.27YY162 pKa = 11.4DD163 pKa = 4.06FGRR166 pKa = 11.84RR167 pKa = 11.84PTTRR171 pKa = 11.84EE172 pKa = 3.46EE173 pKa = 4.06AEE175 pKa = 3.89QLLGIVHH182 pKa = 6.36EE183 pKa = 4.95HH184 pKa = 6.18VCPDD188 pKa = 2.91WW189 pKa = 5.83
Molecular weight: 20.11 kDa
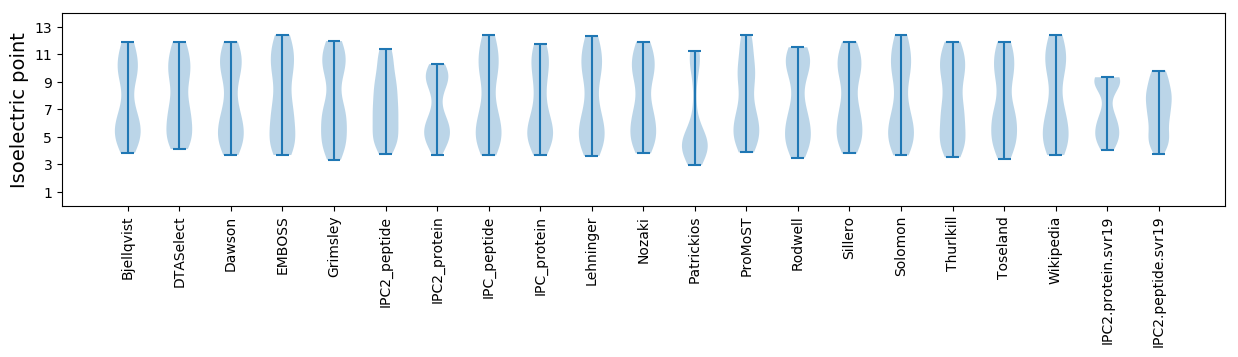
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0R8V9Y8|A0A0R8V9Y8_9CAUD Uncharacterized protein OS=Thermobifida phage P1312 OX=1661715 GN=P1312_070 PE=4 SV=1
MM1 pKa = 7.09AQLVKK6 pKa = 10.26RR7 pKa = 11.84AYY9 pKa = 9.65KK10 pKa = 9.99YY11 pKa = 10.34RR12 pKa = 11.84FYY14 pKa = 10.03PTLEE18 pKa = 3.86QAEE21 pKa = 4.28EE22 pKa = 3.92LLRR25 pKa = 11.84TFGCVRR31 pKa = 11.84FVYY34 pKa = 10.57NKK36 pKa = 10.2ALEE39 pKa = 4.22EE40 pKa = 3.85RR41 pKa = 11.84TRR43 pKa = 11.84AYY45 pKa = 8.09TQEE48 pKa = 3.78GRR50 pKa = 11.84RR51 pKa = 11.84VSYY54 pKa = 11.28VEE56 pKa = 3.74TSAALTQWKK65 pKa = 7.57RR66 pKa = 11.84TPEE69 pKa = 3.81LAFLNEE75 pKa = 4.21VSSVPLQQALRR86 pKa = 11.84HH87 pKa = 5.33LQAAFANFFAKK98 pKa = 9.81RR99 pKa = 11.84AKK101 pKa = 10.71YY102 pKa = 7.94PTFKK106 pKa = 10.55SKK108 pKa = 10.61KK109 pKa = 8.24KK110 pKa = 9.6SRR112 pKa = 11.84ASAEE116 pKa = 4.02YY117 pKa = 9.36TRR119 pKa = 11.84SAFRR123 pKa = 11.84WKK125 pKa = 10.52DD126 pKa = 3.14GRR128 pKa = 11.84LFLAKK133 pKa = 9.3MRR135 pKa = 11.84EE136 pKa = 4.03PLRR139 pKa = 11.84IVWSRR144 pKa = 11.84PLPEE148 pKa = 4.62GAQPSTVTVSRR159 pKa = 11.84DD160 pKa = 2.95AAGRR164 pKa = 11.84WFVSILVEE172 pKa = 4.19EE173 pKa = 5.31KK174 pKa = 10.22IRR176 pKa = 11.84PLPPVEE182 pKa = 4.27SSVGVDD188 pKa = 3.17AGISALVTLSTGEE201 pKa = 4.43KK202 pKa = 7.83ITNPGHH208 pKa = 5.35EE209 pKa = 3.83RR210 pKa = 11.84RR211 pKa = 11.84DD212 pKa = 3.47RR213 pKa = 11.84RR214 pKa = 11.84KK215 pKa = 9.38LAKK218 pKa = 9.84AQRR221 pKa = 11.84ALARR225 pKa = 11.84KK226 pKa = 9.19QKK228 pKa = 10.31GSKK231 pKa = 9.4NRR233 pKa = 11.84EE234 pKa = 3.58KK235 pKa = 11.08ARR237 pKa = 11.84LKK239 pKa = 9.93VARR242 pKa = 11.84IYY244 pKa = 11.39ARR246 pKa = 11.84ITDD249 pKa = 3.45RR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84DD253 pKa = 3.82FLHH256 pKa = 6.95KK257 pKa = 10.01LTTRR261 pKa = 11.84LVRR264 pKa = 11.84EE265 pKa = 4.05NQTVVIEE272 pKa = 4.37DD273 pKa = 3.43LTVRR277 pKa = 11.84NMLRR281 pKa = 11.84NHH283 pKa = 6.08SLARR287 pKa = 11.84AISDD291 pKa = 3.42ASWRR295 pKa = 11.84EE296 pKa = 3.61FRR298 pKa = 11.84SMLEE302 pKa = 4.25YY303 pKa = 10.53KK304 pKa = 10.23CAWYY308 pKa = 9.71DD309 pKa = 3.62RR310 pKa = 11.84EE311 pKa = 4.16LLVVDD316 pKa = 4.28RR317 pKa = 11.84WFPSSKK323 pKa = 10.31LCSACGTLQEE333 pKa = 4.58KK334 pKa = 9.8MPLNVRR340 pKa = 11.84TWQCACGAAHH350 pKa = 7.24DD351 pKa = 4.46RR352 pKa = 11.84DD353 pKa = 3.87VNAARR358 pKa = 11.84NILAAGLAEE367 pKa = 4.06RR368 pKa = 4.91
MM1 pKa = 7.09AQLVKK6 pKa = 10.26RR7 pKa = 11.84AYY9 pKa = 9.65KK10 pKa = 9.99YY11 pKa = 10.34RR12 pKa = 11.84FYY14 pKa = 10.03PTLEE18 pKa = 3.86QAEE21 pKa = 4.28EE22 pKa = 3.92LLRR25 pKa = 11.84TFGCVRR31 pKa = 11.84FVYY34 pKa = 10.57NKK36 pKa = 10.2ALEE39 pKa = 4.22EE40 pKa = 3.85RR41 pKa = 11.84TRR43 pKa = 11.84AYY45 pKa = 8.09TQEE48 pKa = 3.78GRR50 pKa = 11.84RR51 pKa = 11.84VSYY54 pKa = 11.28VEE56 pKa = 3.74TSAALTQWKK65 pKa = 7.57RR66 pKa = 11.84TPEE69 pKa = 3.81LAFLNEE75 pKa = 4.21VSSVPLQQALRR86 pKa = 11.84HH87 pKa = 5.33LQAAFANFFAKK98 pKa = 9.81RR99 pKa = 11.84AKK101 pKa = 10.71YY102 pKa = 7.94PTFKK106 pKa = 10.55SKK108 pKa = 10.61KK109 pKa = 8.24KK110 pKa = 9.6SRR112 pKa = 11.84ASAEE116 pKa = 4.02YY117 pKa = 9.36TRR119 pKa = 11.84SAFRR123 pKa = 11.84WKK125 pKa = 10.52DD126 pKa = 3.14GRR128 pKa = 11.84LFLAKK133 pKa = 9.3MRR135 pKa = 11.84EE136 pKa = 4.03PLRR139 pKa = 11.84IVWSRR144 pKa = 11.84PLPEE148 pKa = 4.62GAQPSTVTVSRR159 pKa = 11.84DD160 pKa = 2.95AAGRR164 pKa = 11.84WFVSILVEE172 pKa = 4.19EE173 pKa = 5.31KK174 pKa = 10.22IRR176 pKa = 11.84PLPPVEE182 pKa = 4.27SSVGVDD188 pKa = 3.17AGISALVTLSTGEE201 pKa = 4.43KK202 pKa = 7.83ITNPGHH208 pKa = 5.35EE209 pKa = 3.83RR210 pKa = 11.84RR211 pKa = 11.84DD212 pKa = 3.47RR213 pKa = 11.84RR214 pKa = 11.84KK215 pKa = 9.38LAKK218 pKa = 9.84AQRR221 pKa = 11.84ALARR225 pKa = 11.84KK226 pKa = 9.19QKK228 pKa = 10.31GSKK231 pKa = 9.4NRR233 pKa = 11.84EE234 pKa = 3.58KK235 pKa = 11.08ARR237 pKa = 11.84LKK239 pKa = 9.93VARR242 pKa = 11.84IYY244 pKa = 11.39ARR246 pKa = 11.84ITDD249 pKa = 3.45RR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84DD253 pKa = 3.82FLHH256 pKa = 6.95KK257 pKa = 10.01LTTRR261 pKa = 11.84LVRR264 pKa = 11.84EE265 pKa = 4.05NQTVVIEE272 pKa = 4.37DD273 pKa = 3.43LTVRR277 pKa = 11.84NMLRR281 pKa = 11.84NHH283 pKa = 6.08SLARR287 pKa = 11.84AISDD291 pKa = 3.42ASWRR295 pKa = 11.84EE296 pKa = 3.61FRR298 pKa = 11.84SMLEE302 pKa = 4.25YY303 pKa = 10.53KK304 pKa = 10.23CAWYY308 pKa = 9.71DD309 pKa = 3.62RR310 pKa = 11.84EE311 pKa = 4.16LLVVDD316 pKa = 4.28RR317 pKa = 11.84WFPSSKK323 pKa = 10.31LCSACGTLQEE333 pKa = 4.58KK334 pKa = 9.8MPLNVRR340 pKa = 11.84TWQCACGAAHH350 pKa = 7.24DD351 pKa = 4.46RR352 pKa = 11.84DD353 pKa = 3.87VNAARR358 pKa = 11.84NILAAGLAEE367 pKa = 4.06RR368 pKa = 4.91
Molecular weight: 42.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
21268 |
75 |
1657 |
276.2 |
30.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.327 ± 0.438 | 0.987 ± 0.131 |
5.694 ± 0.206 | 6.503 ± 0.28 |
2.069 ± 0.231 | 9.216 ± 0.48 |
2.708 ± 0.252 | 3.399 ± 0.178 |
2.285 ± 0.19 | 8.83 ± 0.3 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.655 ± 0.101 | 1.914 ± 0.129 |
6.446 ± 0.316 | 3.682 ± 0.161 |
10.001 ± 0.457 | 5.106 ± 0.266 |
5.99 ± 0.289 | 8.195 ± 0.283 |
1.994 ± 0.162 | 1.998 ± 0.172 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
