
Oxytricha trifallax
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Ciliophora; Intramacronucleata; Spirotrichea; Stichotrichia; Sporadotrichida; Oxytrichidae; Oxytrichinae; Oxytricha
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
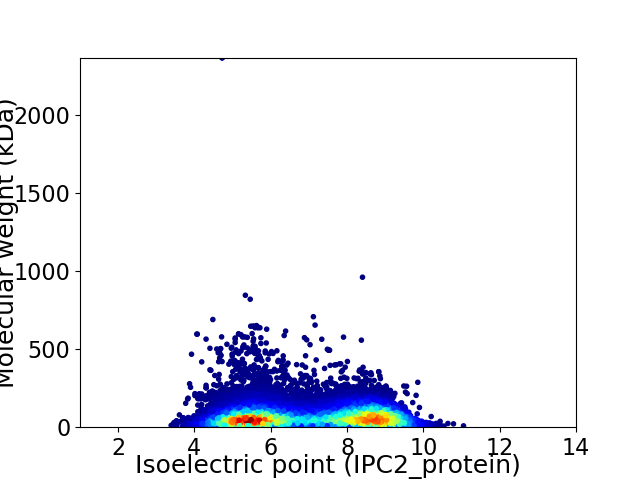
Virtual 2D-PAGE plot for 23559 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J9IE20|J9IE20_9SPIT Uncharacterized protein OS=Oxytricha trifallax OX=1172189 GN=OXYTRI_01707 PE=4 SV=1
MM1 pKa = 7.38LFVYY5 pKa = 10.09KK6 pKa = 10.4NCKK9 pKa = 9.58LLNQVIRR16 pKa = 11.84FSPLFKK22 pKa = 10.36LKK24 pKa = 10.71SIMRR28 pKa = 11.84LAKK31 pKa = 10.12VLALTVVGLIATVGCQDD48 pKa = 3.15QVEE51 pKa = 4.87PGTEE55 pKa = 3.92DD56 pKa = 3.14QQQPEE61 pKa = 4.24IIIPGEE67 pKa = 3.88EE68 pKa = 4.14YY69 pKa = 10.8LRR71 pKa = 11.84AVTGLANSMSSHH83 pKa = 7.46AIPGGVAANITFALQNLSTYY103 pKa = 9.89YY104 pKa = 10.88DD105 pKa = 3.64SFSGFGQLLNQDD117 pKa = 3.75QTVGQYY123 pKa = 11.01LYY125 pKa = 9.66DD126 pKa = 3.58TYY128 pKa = 10.38KK129 pKa = 9.73TQISKK134 pKa = 10.5QDD136 pKa = 3.5DD137 pKa = 3.83AVFGLVDD144 pKa = 4.27RR145 pKa = 11.84NLKK148 pKa = 8.63TLFEE152 pKa = 4.57GQDD155 pKa = 2.98DD156 pKa = 4.87CYY158 pKa = 11.61NDD160 pKa = 3.78FSRR163 pKa = 11.84GIAEE167 pKa = 5.16IFLGWSKK174 pKa = 10.8LWDD177 pKa = 4.91RR178 pKa = 11.84ITGVNRR184 pKa = 11.84YY185 pKa = 8.84GKK187 pKa = 10.52LEE189 pKa = 3.97EE190 pKa = 4.74SKK192 pKa = 11.06DD193 pKa = 3.66FADD196 pKa = 3.83FCVRR200 pKa = 11.84EE201 pKa = 3.88RR202 pKa = 11.84DD203 pKa = 3.81NLSSHH208 pKa = 6.92IDD210 pKa = 4.17DD211 pKa = 4.31IADD214 pKa = 3.41MLLGTEE220 pKa = 4.05QCDD223 pKa = 3.72VLQALYY229 pKa = 10.72NGNVQQNIWTGWRR242 pKa = 11.84DD243 pKa = 3.74NIDD246 pKa = 3.17NNAAYY251 pKa = 10.39LVMYY255 pKa = 7.61FTEE258 pKa = 4.91GVAVFDD264 pKa = 4.2SCHH267 pKa = 5.58EE268 pKa = 4.34VYY270 pKa = 10.34QQLDD274 pKa = 3.1ADD276 pKa = 4.78FAADD280 pKa = 3.38EE281 pKa = 4.43TQQTEE286 pKa = 4.1HH287 pKa = 6.34QAQLDD292 pKa = 3.59QYY294 pKa = 9.7RR295 pKa = 11.84TDD297 pKa = 3.81FQDD300 pKa = 2.64IVNRR304 pKa = 11.84ILEE307 pKa = 4.14FDD309 pKa = 3.44QLALHH314 pKa = 6.6SIEE317 pKa = 4.45LTLPQNLVQLLDD329 pKa = 3.9EE330 pKa = 4.71NQDD333 pKa = 3.21LSLEE337 pKa = 4.24VMGFNVNGFFLSTIYY352 pKa = 9.98WDD354 pKa = 4.17YY355 pKa = 10.99LMQMGAVFSDD365 pKa = 3.31ASEE368 pKa = 3.86DD369 pKa = 3.6AFYY372 pKa = 11.15SEE374 pKa = 5.17CWQCFTVTQGDD385 pKa = 3.98YY386 pKa = 10.6RR387 pKa = 11.84VTVSWLPVEE396 pKa = 4.66GTASVTQEE404 pKa = 3.89TLDD407 pKa = 4.37AIVFDD412 pKa = 4.12EE413 pKa = 4.59TNSKK417 pKa = 10.52EE418 pKa = 4.27DD419 pKa = 3.5LANQVWLGMGDD430 pKa = 3.42EE431 pKa = 4.6CVAGVMIFDD440 pKa = 4.45RR441 pKa = 11.84SIDD444 pKa = 3.67HH445 pKa = 6.22YY446 pKa = 11.82VNTQFTDD453 pKa = 3.31QVVFSQPDD461 pKa = 3.51DD462 pKa = 3.87VDD464 pKa = 4.3IIAWGINEE472 pKa = 4.4LCLGRR477 pKa = 11.84PSLGSTQPVEE487 pKa = 4.06EE488 pKa = 4.35QPTVEE493 pKa = 4.56TPTPADD499 pKa = 3.63TEE501 pKa = 4.2ATTDD505 pKa = 3.58NNEE508 pKa = 4.18EE509 pKa = 4.24SSDD512 pKa = 3.78TQPP515 pKa = 4.01
MM1 pKa = 7.38LFVYY5 pKa = 10.09KK6 pKa = 10.4NCKK9 pKa = 9.58LLNQVIRR16 pKa = 11.84FSPLFKK22 pKa = 10.36LKK24 pKa = 10.71SIMRR28 pKa = 11.84LAKK31 pKa = 10.12VLALTVVGLIATVGCQDD48 pKa = 3.15QVEE51 pKa = 4.87PGTEE55 pKa = 3.92DD56 pKa = 3.14QQQPEE61 pKa = 4.24IIIPGEE67 pKa = 3.88EE68 pKa = 4.14YY69 pKa = 10.8LRR71 pKa = 11.84AVTGLANSMSSHH83 pKa = 7.46AIPGGVAANITFALQNLSTYY103 pKa = 9.89YY104 pKa = 10.88DD105 pKa = 3.64SFSGFGQLLNQDD117 pKa = 3.75QTVGQYY123 pKa = 11.01LYY125 pKa = 9.66DD126 pKa = 3.58TYY128 pKa = 10.38KK129 pKa = 9.73TQISKK134 pKa = 10.5QDD136 pKa = 3.5DD137 pKa = 3.83AVFGLVDD144 pKa = 4.27RR145 pKa = 11.84NLKK148 pKa = 8.63TLFEE152 pKa = 4.57GQDD155 pKa = 2.98DD156 pKa = 4.87CYY158 pKa = 11.61NDD160 pKa = 3.78FSRR163 pKa = 11.84GIAEE167 pKa = 5.16IFLGWSKK174 pKa = 10.8LWDD177 pKa = 4.91RR178 pKa = 11.84ITGVNRR184 pKa = 11.84YY185 pKa = 8.84GKK187 pKa = 10.52LEE189 pKa = 3.97EE190 pKa = 4.74SKK192 pKa = 11.06DD193 pKa = 3.66FADD196 pKa = 3.83FCVRR200 pKa = 11.84EE201 pKa = 3.88RR202 pKa = 11.84DD203 pKa = 3.81NLSSHH208 pKa = 6.92IDD210 pKa = 4.17DD211 pKa = 4.31IADD214 pKa = 3.41MLLGTEE220 pKa = 4.05QCDD223 pKa = 3.72VLQALYY229 pKa = 10.72NGNVQQNIWTGWRR242 pKa = 11.84DD243 pKa = 3.74NIDD246 pKa = 3.17NNAAYY251 pKa = 10.39LVMYY255 pKa = 7.61FTEE258 pKa = 4.91GVAVFDD264 pKa = 4.2SCHH267 pKa = 5.58EE268 pKa = 4.34VYY270 pKa = 10.34QQLDD274 pKa = 3.1ADD276 pKa = 4.78FAADD280 pKa = 3.38EE281 pKa = 4.43TQQTEE286 pKa = 4.1HH287 pKa = 6.34QAQLDD292 pKa = 3.59QYY294 pKa = 9.7RR295 pKa = 11.84TDD297 pKa = 3.81FQDD300 pKa = 2.64IVNRR304 pKa = 11.84ILEE307 pKa = 4.14FDD309 pKa = 3.44QLALHH314 pKa = 6.6SIEE317 pKa = 4.45LTLPQNLVQLLDD329 pKa = 3.9EE330 pKa = 4.71NQDD333 pKa = 3.21LSLEE337 pKa = 4.24VMGFNVNGFFLSTIYY352 pKa = 9.98WDD354 pKa = 4.17YY355 pKa = 10.99LMQMGAVFSDD365 pKa = 3.31ASEE368 pKa = 3.86DD369 pKa = 3.6AFYY372 pKa = 11.15SEE374 pKa = 5.17CWQCFTVTQGDD385 pKa = 3.98YY386 pKa = 10.6RR387 pKa = 11.84VTVSWLPVEE396 pKa = 4.66GTASVTQEE404 pKa = 3.89TLDD407 pKa = 4.37AIVFDD412 pKa = 4.12EE413 pKa = 4.59TNSKK417 pKa = 10.52EE418 pKa = 4.27DD419 pKa = 3.5LANQVWLGMGDD430 pKa = 3.42EE431 pKa = 4.6CVAGVMIFDD440 pKa = 4.45RR441 pKa = 11.84SIDD444 pKa = 3.67HH445 pKa = 6.22YY446 pKa = 11.82VNTQFTDD453 pKa = 3.31QVVFSQPDD461 pKa = 3.51DD462 pKa = 3.87VDD464 pKa = 4.3IIAWGINEE472 pKa = 4.4LCLGRR477 pKa = 11.84PSLGSTQPVEE487 pKa = 4.06EE488 pKa = 4.35QPTVEE493 pKa = 4.56TPTPADD499 pKa = 3.63TEE501 pKa = 4.2ATTDD505 pKa = 3.58NNEE508 pKa = 4.18EE509 pKa = 4.24SSDD512 pKa = 3.78TQPP515 pKa = 4.01
Molecular weight: 58.04 kDa
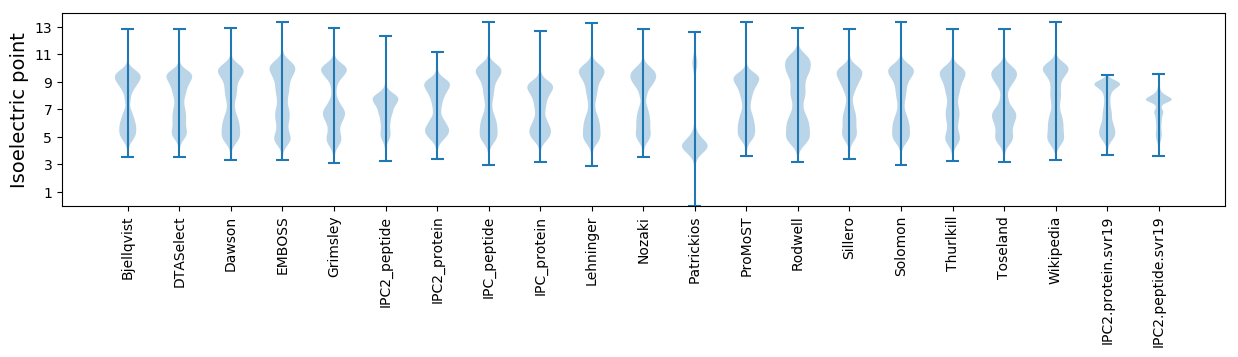
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J9HMT3|J9HMT3_9SPIT Serine/threonine protein kinase OS=Oxytricha trifallax OX=1172189 GN=OXYTRI_14793 PE=4 SV=1
MM1 pKa = 7.52ARR3 pKa = 11.84TAAPKK8 pKa = 10.04RR9 pKa = 11.84VAKK12 pKa = 10.51RR13 pKa = 11.84PTKK16 pKa = 10.27KK17 pKa = 10.47AGSKK21 pKa = 10.11KK22 pKa = 8.73IAKK25 pKa = 9.98RR26 pKa = 11.84PVAKK30 pKa = 10.33KK31 pKa = 10.37AGAKK35 pKa = 9.23RR36 pKa = 11.84VAKK39 pKa = 10.28KK40 pKa = 8.88ATPVAAAPVVAGSTSATPQKK60 pKa = 10.72AAAKK64 pKa = 9.94KK65 pKa = 9.12SAKK68 pKa = 9.72KK69 pKa = 9.58AAKK72 pKa = 10.08RR73 pKa = 11.84PAKK76 pKa = 9.97KK77 pKa = 9.91AAPKK81 pKa = 10.26RR82 pKa = 11.84RR83 pKa = 3.57
MM1 pKa = 7.52ARR3 pKa = 11.84TAAPKK8 pKa = 10.04RR9 pKa = 11.84VAKK12 pKa = 10.51RR13 pKa = 11.84PTKK16 pKa = 10.27KK17 pKa = 10.47AGSKK21 pKa = 10.11KK22 pKa = 8.73IAKK25 pKa = 9.98RR26 pKa = 11.84PVAKK30 pKa = 10.33KK31 pKa = 10.37AGAKK35 pKa = 9.23RR36 pKa = 11.84VAKK39 pKa = 10.28KK40 pKa = 8.88ATPVAAAPVVAGSTSATPQKK60 pKa = 10.72AAAKK64 pKa = 9.94KK65 pKa = 9.12SAKK68 pKa = 9.72KK69 pKa = 9.58AAKK72 pKa = 10.08RR73 pKa = 11.84PAKK76 pKa = 9.97KK77 pKa = 9.91AAPKK81 pKa = 10.26RR82 pKa = 11.84RR83 pKa = 3.57
Molecular weight: 8.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
15273496 |
49 |
21680 |
648.3 |
74.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.901 ± 0.011 | 1.15 ± 0.009 |
5.628 ± 0.009 | 6.216 ± 0.02 |
4.569 ± 0.011 | 4.179 ± 0.016 |
1.915 ± 0.007 | 7.208 ± 0.014 |
8.4 ± 0.022 | 8.675 ± 0.013 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.443 ± 0.006 | 7.941 ± 0.017 |
3.11 ± 0.012 | 8.963 ± 0.022 |
3.851 ± 0.012 | 8.233 ± 0.016 |
4.91 ± 0.021 | 4.407 ± 0.01 |
0.579 ± 0.004 | 3.72 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
