
Teredinibacter turnerae (strain ATCC 39867 / T7901)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Cellvibrionaceae; Teredinibacter; Teredinibacter turnerae
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
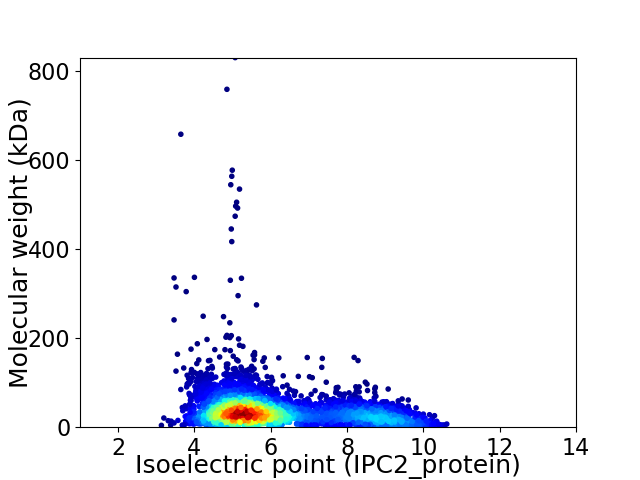
Virtual 2D-PAGE plot for 4249 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C5BKC5|C5BKC5_TERTT Uncharacterized protein OS=Teredinibacter turnerae (strain ATCC 39867 / T7901) OX=377629 GN=TERTU_2389 PE=4 SV=1
MM1 pKa = 7.48LPKK4 pKa = 10.04TYY6 pKa = 10.55LIGVAFGLLAVTAEE20 pKa = 4.26SATVCDD26 pKa = 3.97TVDD29 pKa = 3.37NDD31 pKa = 4.09GDD33 pKa = 4.0PFDD36 pKa = 3.79VGTYY40 pKa = 7.88GACGASSSDD49 pKa = 3.94LSGVWMIVSDD59 pKa = 4.28YY60 pKa = 11.14NLEE63 pKa = 3.9TSYY66 pKa = 11.37FGMTWNEE73 pKa = 3.83SRR75 pKa = 11.84RR76 pKa = 11.84QRR78 pKa = 11.84QTVQILDD85 pKa = 3.93NQDD88 pKa = 2.75GTITVSDD95 pKa = 4.45CGFLGYY101 pKa = 10.39GYY103 pKa = 10.07EE104 pKa = 4.36YY105 pKa = 10.67TLSTSANEE113 pKa = 3.85LTLVGDD119 pKa = 3.67QVVDD123 pKa = 4.52FSLQIVNNTQLSGSVVSRR141 pKa = 11.84ASVWGTSIDD150 pKa = 3.93VISNYY155 pKa = 10.68AEE157 pKa = 3.87ATKK160 pKa = 10.71LRR162 pKa = 11.84DD163 pKa = 3.12LTAPVVTLGDD173 pKa = 3.73LSSEE177 pKa = 4.09YY178 pKa = 11.3GPVDD182 pKa = 3.45KK183 pKa = 11.09KK184 pKa = 11.56KK185 pKa = 10.08MLNNWQIACFKK196 pKa = 8.95NTAYY200 pKa = 10.52DD201 pKa = 3.27IEE203 pKa = 4.4YY204 pKa = 8.85TYY206 pKa = 11.53LDD208 pKa = 3.66GSTEE212 pKa = 3.74RR213 pKa = 11.84RR214 pKa = 11.84VYY216 pKa = 11.12EE217 pKa = 4.01NTDD220 pKa = 3.12LYY222 pKa = 11.26AAEE225 pKa = 4.25SVEE228 pKa = 4.18NGIAGDD234 pKa = 3.89ALVLFSISEE243 pKa = 3.95PTLNEE248 pKa = 3.53EE249 pKa = 4.87LRR251 pKa = 11.84AEE253 pKa = 4.18FNLAEE258 pKa = 4.41RR259 pKa = 11.84NDD261 pKa = 3.91VNGEE265 pKa = 3.91ADD267 pKa = 3.79NVNIDD272 pKa = 3.56YY273 pKa = 11.13VADD276 pKa = 4.16SNSHH280 pKa = 5.99LSGIASIQDD289 pKa = 3.07EE290 pKa = 4.21DD291 pKa = 4.09RR292 pKa = 11.84ALVSADD298 pKa = 2.58IKK300 pKa = 11.41FNFSLL305 pKa = 4.22
MM1 pKa = 7.48LPKK4 pKa = 10.04TYY6 pKa = 10.55LIGVAFGLLAVTAEE20 pKa = 4.26SATVCDD26 pKa = 3.97TVDD29 pKa = 3.37NDD31 pKa = 4.09GDD33 pKa = 4.0PFDD36 pKa = 3.79VGTYY40 pKa = 7.88GACGASSSDD49 pKa = 3.94LSGVWMIVSDD59 pKa = 4.28YY60 pKa = 11.14NLEE63 pKa = 3.9TSYY66 pKa = 11.37FGMTWNEE73 pKa = 3.83SRR75 pKa = 11.84RR76 pKa = 11.84QRR78 pKa = 11.84QTVQILDD85 pKa = 3.93NQDD88 pKa = 2.75GTITVSDD95 pKa = 4.45CGFLGYY101 pKa = 10.39GYY103 pKa = 10.07EE104 pKa = 4.36YY105 pKa = 10.67TLSTSANEE113 pKa = 3.85LTLVGDD119 pKa = 3.67QVVDD123 pKa = 4.52FSLQIVNNTQLSGSVVSRR141 pKa = 11.84ASVWGTSIDD150 pKa = 3.93VISNYY155 pKa = 10.68AEE157 pKa = 3.87ATKK160 pKa = 10.71LRR162 pKa = 11.84DD163 pKa = 3.12LTAPVVTLGDD173 pKa = 3.73LSSEE177 pKa = 4.09YY178 pKa = 11.3GPVDD182 pKa = 3.45KK183 pKa = 11.09KK184 pKa = 11.56KK185 pKa = 10.08MLNNWQIACFKK196 pKa = 8.95NTAYY200 pKa = 10.52DD201 pKa = 3.27IEE203 pKa = 4.4YY204 pKa = 8.85TYY206 pKa = 11.53LDD208 pKa = 3.66GSTEE212 pKa = 3.74RR213 pKa = 11.84RR214 pKa = 11.84VYY216 pKa = 11.12EE217 pKa = 4.01NTDD220 pKa = 3.12LYY222 pKa = 11.26AAEE225 pKa = 4.25SVEE228 pKa = 4.18NGIAGDD234 pKa = 3.89ALVLFSISEE243 pKa = 3.95PTLNEE248 pKa = 3.53EE249 pKa = 4.87LRR251 pKa = 11.84AEE253 pKa = 4.18FNLAEE258 pKa = 4.41RR259 pKa = 11.84NDD261 pKa = 3.91VNGEE265 pKa = 3.91ADD267 pKa = 3.79NVNIDD272 pKa = 3.56YY273 pKa = 11.13VADD276 pKa = 4.16SNSHH280 pKa = 5.99LSGIASIQDD289 pKa = 3.07EE290 pKa = 4.21DD291 pKa = 4.09RR292 pKa = 11.84ALVSADD298 pKa = 2.58IKK300 pKa = 11.41FNFSLL305 pKa = 4.22
Molecular weight: 33.25 kDa
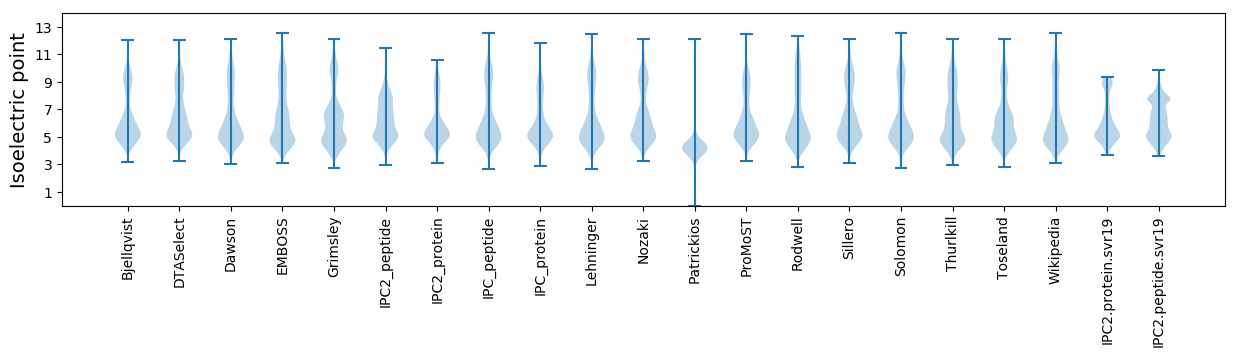
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C5BJ77|C5BJ77_TERTT Uncharacterized protein OS=Teredinibacter turnerae (strain ATCC 39867 / T7901) OX=377629 GN=TERTU_4491 PE=4 SV=1
MM1 pKa = 7.73HH2 pKa = 7.28NNQHH6 pKa = 5.34QAQPLRR12 pKa = 11.84GLDD15 pKa = 3.4LHH17 pKa = 6.91FGASRR22 pKa = 11.84LYY24 pKa = 10.51CGPCAGRR31 pKa = 11.84YY32 pKa = 7.57KK33 pKa = 10.62ASKK36 pKa = 10.38ASILKK41 pKa = 10.14RR42 pKa = 11.84YY43 pKa = 9.88VLVKK47 pKa = 9.19RR48 pKa = 11.84CRR50 pKa = 11.84VVRR53 pKa = 11.84DD54 pKa = 3.48LVAA57 pKa = 5.17
MM1 pKa = 7.73HH2 pKa = 7.28NNQHH6 pKa = 5.34QAQPLRR12 pKa = 11.84GLDD15 pKa = 3.4LHH17 pKa = 6.91FGASRR22 pKa = 11.84LYY24 pKa = 10.51CGPCAGRR31 pKa = 11.84YY32 pKa = 7.57KK33 pKa = 10.62ASKK36 pKa = 10.38ASILKK41 pKa = 10.14RR42 pKa = 11.84YY43 pKa = 9.88VLVKK47 pKa = 9.19RR48 pKa = 11.84CRR50 pKa = 11.84VVRR53 pKa = 11.84DD54 pKa = 3.48LVAA57 pKa = 5.17
Molecular weight: 6.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1491791 |
37 |
7446 |
351.1 |
38.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.759 ± 0.046 | 1.104 ± 0.015 |
5.747 ± 0.042 | 6.066 ± 0.037 |
4.055 ± 0.025 | 7.201 ± 0.057 |
2.186 ± 0.02 | 5.505 ± 0.028 |
4.158 ± 0.037 | 10.283 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.226 ± 0.019 | 4.092 ± 0.034 |
4.315 ± 0.029 | 4.197 ± 0.029 |
5.281 ± 0.036 | 6.902 ± 0.055 |
5.397 ± 0.036 | 7.088 ± 0.03 |
1.361 ± 0.017 | 3.076 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
