
Vittaforma corneae (strain ATCC 50505) (Microsporidian parasite) (Nosema corneum)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Microsporidia; Apansporoblastina; Nosematidae; Vittaforma; Vittaforma corneae
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
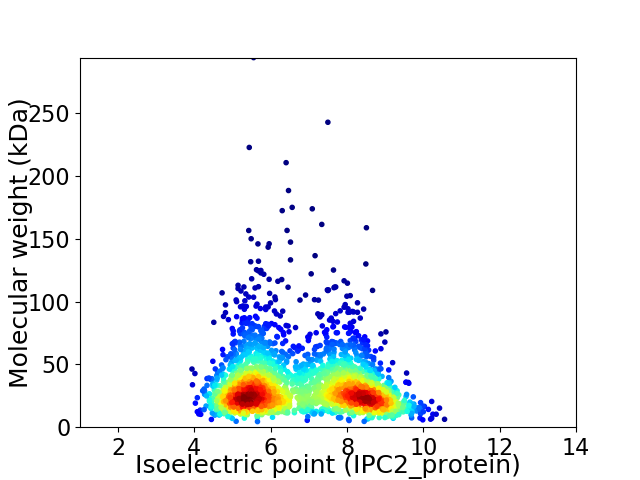
Virtual 2D-PAGE plot for 2236 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L2GK29|L2GK29_VITCO Uncharacterized protein (Fragment) OS=Vittaforma corneae (strain ATCC 50505) OX=993615 GN=VICG_01727 PE=4 SV=1
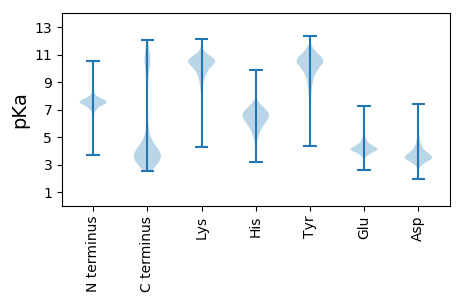
MM1 pKa = 7.85PKK3 pKa = 10.01QSTEE7 pKa = 3.59NRR9 pKa = 11.84QDD11 pKa = 3.2SNMIFIDD18 pKa = 4.13DD19 pKa = 4.04PEE21 pKa = 5.19LNEE24 pKa = 4.64EE25 pKa = 4.21YY26 pKa = 10.11PIDD29 pKa = 3.87FTDD32 pKa = 3.68VNSDD36 pKa = 3.34SAGMITIQSSTEE48 pKa = 3.89DD49 pKa = 3.93EE50 pKa = 4.17NTVIVDD56 pKa = 3.67EE57 pKa = 4.93GSLIEE62 pKa = 5.64DD63 pKa = 4.17SLSHH67 pKa = 6.4SNPVFNSSFEE77 pKa = 4.39EE78 pKa = 4.07YY79 pKa = 9.73RR80 pKa = 11.84SSYY83 pKa = 9.54EE84 pKa = 4.01GQVDD88 pKa = 4.31EE89 pKa = 4.85YY90 pKa = 11.59SSFDD94 pKa = 3.44SSGGTLYY101 pKa = 10.96NEE103 pKa = 4.39EE104 pKa = 4.41YY105 pKa = 10.8SFEE108 pKa = 5.04DD109 pKa = 4.75EE110 pKa = 5.61FDD112 pKa = 3.95DD113 pKa = 6.71DD114 pKa = 4.4YY115 pKa = 12.03DD116 pKa = 3.78EE117 pKa = 6.06FEE119 pKa = 5.09DD120 pKa = 4.54EE121 pKa = 4.66EE122 pKa = 4.5EE123 pKa = 3.83EE124 pKa = 4.46SYY126 pKa = 11.48YY127 pKa = 10.95FSDD130 pKa = 5.48DD131 pKa = 4.14LDD133 pKa = 3.82ATLYY137 pKa = 10.86DD138 pKa = 3.73YY139 pKa = 11.61DD140 pKa = 4.48EE141 pKa = 5.26DD142 pKa = 4.27SEE144 pKa = 4.85YY145 pKa = 11.2SDD147 pKa = 3.09EE148 pKa = 4.64VYY150 pKa = 9.85WEE152 pKa = 4.11EE153 pKa = 3.7TSQIVTGMRR162 pKa = 11.84RR163 pKa = 11.84GEE165 pKa = 3.84LSRR168 pKa = 11.84FPEE171 pKa = 4.44NGNSTGADD179 pKa = 3.1RR180 pKa = 11.84NEE182 pKa = 3.92RR183 pKa = 11.84LSAEE187 pKa = 4.27SEE189 pKa = 4.05PDD191 pKa = 3.08EE192 pKa = 5.29HH193 pKa = 7.45EE194 pKa = 4.93LIILEE199 pKa = 4.52SRR201 pKa = 11.84NSDD204 pKa = 3.18QLYY207 pKa = 9.94PRR209 pKa = 11.84VTYY212 pKa = 10.29EE213 pKa = 4.91LITMILVRR221 pKa = 11.84MYY223 pKa = 9.3MAYY226 pKa = 10.63NLGIGEE232 pKa = 4.46YY233 pKa = 9.45YY234 pKa = 10.48SIYY237 pKa = 10.68EE238 pKa = 4.04FDD240 pKa = 3.77SYY242 pKa = 11.35PYY244 pKa = 10.7NDD246 pKa = 3.44VNIADD251 pKa = 5.17KK252 pKa = 10.94IRR254 pKa = 11.84DD255 pKa = 3.86FTPDD259 pKa = 3.01EE260 pKa = 4.17LSFLKK265 pKa = 10.81SRR267 pKa = 11.84VSSILMNDD275 pKa = 2.67IRR277 pKa = 11.84DD278 pKa = 3.81VSSVQALISLISIDD292 pKa = 5.23DD293 pKa = 3.31IDD295 pKa = 4.86NISFSSFKK303 pKa = 9.34KK304 pKa = 9.11TKK306 pKa = 10.05FKK308 pKa = 10.73STMFKK313 pKa = 11.35DD314 pKa = 3.94MINVDD319 pKa = 3.79GKK321 pKa = 11.2SCGICYY327 pKa = 10.15CDD329 pKa = 4.02FRR331 pKa = 11.84PSHH334 pKa = 6.62KK335 pKa = 9.66CTSLQCLHH343 pKa = 6.41TFHH346 pKa = 6.76TKK348 pKa = 10.5CIKK351 pKa = 10.28KK352 pKa = 8.57WVAVSWCCPLCRR364 pKa = 11.84RR365 pKa = 11.84KK366 pKa = 10.67DD367 pKa = 3.3IGG369 pKa = 3.52
MM1 pKa = 7.85PKK3 pKa = 10.01QSTEE7 pKa = 3.59NRR9 pKa = 11.84QDD11 pKa = 3.2SNMIFIDD18 pKa = 4.13DD19 pKa = 4.04PEE21 pKa = 5.19LNEE24 pKa = 4.64EE25 pKa = 4.21YY26 pKa = 10.11PIDD29 pKa = 3.87FTDD32 pKa = 3.68VNSDD36 pKa = 3.34SAGMITIQSSTEE48 pKa = 3.89DD49 pKa = 3.93EE50 pKa = 4.17NTVIVDD56 pKa = 3.67EE57 pKa = 4.93GSLIEE62 pKa = 5.64DD63 pKa = 4.17SLSHH67 pKa = 6.4SNPVFNSSFEE77 pKa = 4.39EE78 pKa = 4.07YY79 pKa = 9.73RR80 pKa = 11.84SSYY83 pKa = 9.54EE84 pKa = 4.01GQVDD88 pKa = 4.31EE89 pKa = 4.85YY90 pKa = 11.59SSFDD94 pKa = 3.44SSGGTLYY101 pKa = 10.96NEE103 pKa = 4.39EE104 pKa = 4.41YY105 pKa = 10.8SFEE108 pKa = 5.04DD109 pKa = 4.75EE110 pKa = 5.61FDD112 pKa = 3.95DD113 pKa = 6.71DD114 pKa = 4.4YY115 pKa = 12.03DD116 pKa = 3.78EE117 pKa = 6.06FEE119 pKa = 5.09DD120 pKa = 4.54EE121 pKa = 4.66EE122 pKa = 4.5EE123 pKa = 3.83EE124 pKa = 4.46SYY126 pKa = 11.48YY127 pKa = 10.95FSDD130 pKa = 5.48DD131 pKa = 4.14LDD133 pKa = 3.82ATLYY137 pKa = 10.86DD138 pKa = 3.73YY139 pKa = 11.61DD140 pKa = 4.48EE141 pKa = 5.26DD142 pKa = 4.27SEE144 pKa = 4.85YY145 pKa = 11.2SDD147 pKa = 3.09EE148 pKa = 4.64VYY150 pKa = 9.85WEE152 pKa = 4.11EE153 pKa = 3.7TSQIVTGMRR162 pKa = 11.84RR163 pKa = 11.84GEE165 pKa = 3.84LSRR168 pKa = 11.84FPEE171 pKa = 4.44NGNSTGADD179 pKa = 3.1RR180 pKa = 11.84NEE182 pKa = 3.92RR183 pKa = 11.84LSAEE187 pKa = 4.27SEE189 pKa = 4.05PDD191 pKa = 3.08EE192 pKa = 5.29HH193 pKa = 7.45EE194 pKa = 4.93LIILEE199 pKa = 4.52SRR201 pKa = 11.84NSDD204 pKa = 3.18QLYY207 pKa = 9.94PRR209 pKa = 11.84VTYY212 pKa = 10.29EE213 pKa = 4.91LITMILVRR221 pKa = 11.84MYY223 pKa = 9.3MAYY226 pKa = 10.63NLGIGEE232 pKa = 4.46YY233 pKa = 9.45YY234 pKa = 10.48SIYY237 pKa = 10.68EE238 pKa = 4.04FDD240 pKa = 3.77SYY242 pKa = 11.35PYY244 pKa = 10.7NDD246 pKa = 3.44VNIADD251 pKa = 5.17KK252 pKa = 10.94IRR254 pKa = 11.84DD255 pKa = 3.86FTPDD259 pKa = 3.01EE260 pKa = 4.17LSFLKK265 pKa = 10.81SRR267 pKa = 11.84VSSILMNDD275 pKa = 2.67IRR277 pKa = 11.84DD278 pKa = 3.81VSSVQALISLISIDD292 pKa = 5.23DD293 pKa = 3.31IDD295 pKa = 4.86NISFSSFKK303 pKa = 9.34KK304 pKa = 9.11TKK306 pKa = 10.05FKK308 pKa = 10.73STMFKK313 pKa = 11.35DD314 pKa = 3.94MINVDD319 pKa = 3.79GKK321 pKa = 11.2SCGICYY327 pKa = 10.15CDD329 pKa = 4.02FRR331 pKa = 11.84PSHH334 pKa = 6.62KK335 pKa = 9.66CTSLQCLHH343 pKa = 6.41TFHH346 pKa = 6.76TKK348 pKa = 10.5CIKK351 pKa = 10.28KK352 pKa = 8.57WVAVSWCCPLCRR364 pKa = 11.84RR365 pKa = 11.84KK366 pKa = 10.67DD367 pKa = 3.3IGG369 pKa = 3.52
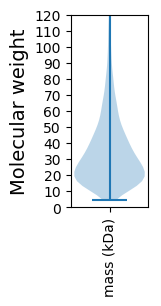
Molecular weight: 42.74 kDa
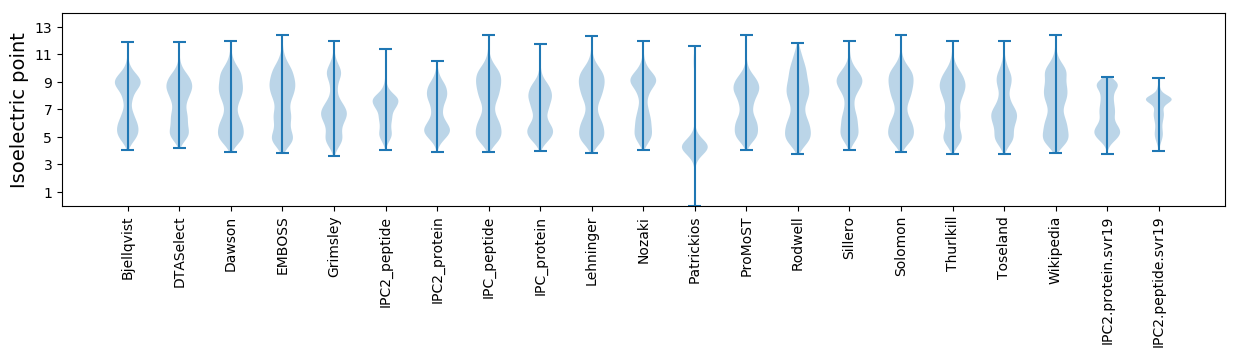
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L2GQ25|L2GQ25_VITCO Glutamyl-tRNA synthetase OS=Vittaforma corneae (strain ATCC 50505) OX=993615 GN=VICG_00261 PE=3 SV=1
MM1 pKa = 7.75AGRR4 pKa = 11.84GKK6 pKa = 10.15GAKK9 pKa = 8.61GAKK12 pKa = 9.2GVAKK16 pKa = 10.26RR17 pKa = 11.84HH18 pKa = 5.75RR19 pKa = 11.84KK20 pKa = 8.74QPSSSSKK27 pKa = 10.05ISGPAIRR34 pKa = 11.84RR35 pKa = 11.84LARR38 pKa = 11.84RR39 pKa = 11.84AGVPRR44 pKa = 11.84VGAGCFPEE52 pKa = 4.66VNDD55 pKa = 4.25AAVNYY60 pKa = 7.52LTTLLDD66 pKa = 3.32SCRR69 pKa = 11.84IYY71 pKa = 10.76CSHH74 pKa = 7.21SKK76 pKa = 10.5RR77 pKa = 11.84KK78 pKa = 8.07TINCQDD84 pKa = 3.25VIYY87 pKa = 10.45ALKK90 pKa = 10.67RR91 pKa = 11.84LGTKK95 pKa = 10.53YY96 pKa = 10.04MGFF99 pKa = 3.7
MM1 pKa = 7.75AGRR4 pKa = 11.84GKK6 pKa = 10.15GAKK9 pKa = 8.61GAKK12 pKa = 9.2GVAKK16 pKa = 10.26RR17 pKa = 11.84HH18 pKa = 5.75RR19 pKa = 11.84KK20 pKa = 8.74QPSSSSKK27 pKa = 10.05ISGPAIRR34 pKa = 11.84RR35 pKa = 11.84LARR38 pKa = 11.84RR39 pKa = 11.84AGVPRR44 pKa = 11.84VGAGCFPEE52 pKa = 4.66VNDD55 pKa = 4.25AAVNYY60 pKa = 7.52LTTLLDD66 pKa = 3.32SCRR69 pKa = 11.84IYY71 pKa = 10.76CSHH74 pKa = 7.21SKK76 pKa = 10.5RR77 pKa = 11.84KK78 pKa = 8.07TINCQDD84 pKa = 3.25VIYY87 pKa = 10.45ALKK90 pKa = 10.67RR91 pKa = 11.84LGTKK95 pKa = 10.53YY96 pKa = 10.04MGFF99 pKa = 3.7
Molecular weight: 10.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
714266 |
39 |
2539 |
319.4 |
36.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.547 ± 0.036 | 2.036 ± 0.024 |
5.507 ± 0.037 | 7.179 ± 0.05 |
5.647 ± 0.041 | 4.341 ± 0.038 |
1.913 ± 0.019 | 8.174 ± 0.043 |
8.357 ± 0.049 | 9.972 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.316 ± 0.018 | 5.913 ± 0.042 |
3.142 ± 0.034 | 3.033 ± 0.024 |
4.407 ± 0.037 | 8.351 ± 0.051 |
4.803 ± 0.024 | 5.854 ± 0.04 |
0.574 ± 0.011 | 3.933 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
