
Methyloligella halotolerans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Hyphomicrobiaceae; Methyloligella
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
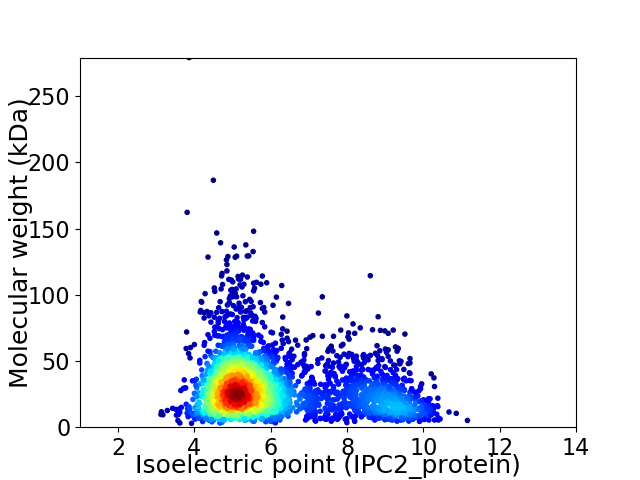
Virtual 2D-PAGE plot for 3174 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E2RZ83|A0A1E2RZ83_9RHIZ NAD(P) transhydrogenase subunit beta OS=Methyloligella halotolerans OX=1177755 GN=A7A08_01558 PE=4 SV=1
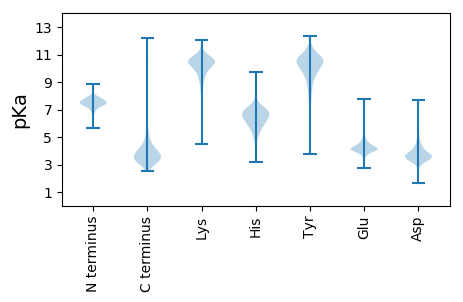
MM1 pKa = 7.01YY2 pKa = 10.63ADD4 pKa = 3.88LGFRR8 pKa = 11.84NSKK11 pKa = 11.11GEE13 pKa = 3.71VHH15 pKa = 6.64VNVTAADD22 pKa = 3.59TSFGVTASAPVQLLALDD39 pKa = 4.05RR40 pKa = 11.84EE41 pKa = 4.63RR42 pKa = 11.84TFTSPQTTDD51 pKa = 2.9NKK53 pKa = 10.47MIMPSVNASYY63 pKa = 11.05NVNDD67 pKa = 4.59NITLSGVAYY76 pKa = 9.74YY77 pKa = 10.57RR78 pKa = 11.84RR79 pKa = 11.84FEE81 pKa = 4.0QSHH84 pKa = 7.07DD85 pKa = 3.69DD86 pKa = 3.96GNISEE91 pKa = 5.88AEE93 pKa = 4.07PCDD96 pKa = 4.17DD97 pKa = 5.33NPAILCLEE105 pKa = 4.38GEE107 pKa = 4.43EE108 pKa = 4.33ATDD111 pKa = 3.44ISGNLIPTPDD121 pKa = 4.05GEE123 pKa = 4.66LGSIDD128 pKa = 3.72STGQTADD135 pKa = 2.63SWGTAVQMANDD146 pKa = 3.81ADD148 pKa = 4.52LYY150 pKa = 10.19GHH152 pKa = 6.9GNHH155 pKa = 6.99FIIGASYY162 pKa = 11.43DD163 pKa = 3.41HH164 pKa = 6.6GQVTFNAQSEE174 pKa = 4.38LAQFQPRR181 pKa = 11.84FVVTPTGTTFGGPDD195 pKa = 3.18EE196 pKa = 4.51VAPKK200 pKa = 10.77LIDD203 pKa = 3.5TEE205 pKa = 4.1NTYY208 pKa = 11.11VGLYY212 pKa = 10.17ISDD215 pKa = 3.69TYY217 pKa = 11.1EE218 pKa = 3.88VNDD221 pKa = 4.19RR222 pKa = 11.84LAVTLGGRR230 pKa = 11.84YY231 pKa = 9.39NYY233 pKa = 11.13ASIQLTDD240 pKa = 3.85LTGEE244 pKa = 4.25DD245 pKa = 3.65PALNGTNTYY254 pKa = 10.41EE255 pKa = 4.28RR256 pKa = 11.84FNPAAGLTYY265 pKa = 10.61QLTSTISAYY274 pKa = 10.33GGYY277 pKa = 9.49SEE279 pKa = 5.94ANRR282 pKa = 11.84APVPAEE288 pKa = 3.84LACADD293 pKa = 4.12PEE295 pKa = 4.4RR296 pKa = 11.84PCLIEE301 pKa = 4.31SFLVADD307 pKa = 4.52PPLEE311 pKa = 3.9QVVSNTWEE319 pKa = 3.92LGLRR323 pKa = 11.84GEE325 pKa = 4.62DD326 pKa = 3.3TAADD330 pKa = 4.18GSSRR334 pKa = 11.84WTWNAGLFRR343 pKa = 11.84TYY345 pKa = 10.72TDD347 pKa = 3.85DD348 pKa = 6.22DD349 pKa = 4.68IITVVTNSTRR359 pKa = 11.84GYY361 pKa = 8.42FQNGADD367 pKa = 3.82ILRR370 pKa = 11.84QGIEE374 pKa = 3.9AGLTYY379 pKa = 9.02QTQKK383 pKa = 7.94WQAYY387 pKa = 9.2ANYY390 pKa = 9.91AYY392 pKa = 9.75IDD394 pKa = 3.32ATYY397 pKa = 11.08QNDD400 pKa = 4.99LILNAPDD407 pKa = 3.54NPSARR412 pKa = 11.84EE413 pKa = 4.15SICDD417 pKa = 3.22ASDD420 pKa = 3.58FEE422 pKa = 4.5EE423 pKa = 4.71EE424 pKa = 4.18EE425 pKa = 4.46EE426 pKa = 4.12EE427 pKa = 5.34GEE429 pKa = 4.94DD430 pKa = 3.96EE431 pKa = 6.23GEE433 pKa = 4.11LACIAVRR440 pKa = 11.84PGDD443 pKa = 4.13QLPGIPPHH451 pKa = 6.46RR452 pKa = 11.84FKK454 pKa = 11.36AGMDD458 pKa = 3.24YY459 pKa = 10.71WVTDD463 pKa = 2.81KK464 pKa = 11.07WKK466 pKa = 10.53IGGDD470 pKa = 3.97LIGVSSQYY478 pKa = 10.82FFGDD482 pKa = 3.47EE483 pKa = 4.55ANQQPQLDD491 pKa = 4.74GYY493 pKa = 9.83WRR495 pKa = 11.84VDD497 pKa = 3.03LHH499 pKa = 6.23TSYY502 pKa = 11.05QITPRR507 pKa = 11.84IQLFALANNVFDD519 pKa = 4.29KK520 pKa = 10.79EE521 pKa = 4.29YY522 pKa = 10.89GVFGTFFNLAGANEE536 pKa = 4.2GAEE539 pKa = 4.19ADD541 pKa = 3.99PDD543 pKa = 3.68LGEE546 pKa = 5.77DD547 pKa = 4.16FFTNPRR553 pKa = 11.84TITPAPPAVIYY564 pKa = 10.53GGAKK568 pKa = 8.34IALWW572 pKa = 3.66
MM1 pKa = 7.01YY2 pKa = 10.63ADD4 pKa = 3.88LGFRR8 pKa = 11.84NSKK11 pKa = 11.11GEE13 pKa = 3.71VHH15 pKa = 6.64VNVTAADD22 pKa = 3.59TSFGVTASAPVQLLALDD39 pKa = 4.05RR40 pKa = 11.84EE41 pKa = 4.63RR42 pKa = 11.84TFTSPQTTDD51 pKa = 2.9NKK53 pKa = 10.47MIMPSVNASYY63 pKa = 11.05NVNDD67 pKa = 4.59NITLSGVAYY76 pKa = 9.74YY77 pKa = 10.57RR78 pKa = 11.84RR79 pKa = 11.84FEE81 pKa = 4.0QSHH84 pKa = 7.07DD85 pKa = 3.69DD86 pKa = 3.96GNISEE91 pKa = 5.88AEE93 pKa = 4.07PCDD96 pKa = 4.17DD97 pKa = 5.33NPAILCLEE105 pKa = 4.38GEE107 pKa = 4.43EE108 pKa = 4.33ATDD111 pKa = 3.44ISGNLIPTPDD121 pKa = 4.05GEE123 pKa = 4.66LGSIDD128 pKa = 3.72STGQTADD135 pKa = 2.63SWGTAVQMANDD146 pKa = 3.81ADD148 pKa = 4.52LYY150 pKa = 10.19GHH152 pKa = 6.9GNHH155 pKa = 6.99FIIGASYY162 pKa = 11.43DD163 pKa = 3.41HH164 pKa = 6.6GQVTFNAQSEE174 pKa = 4.38LAQFQPRR181 pKa = 11.84FVVTPTGTTFGGPDD195 pKa = 3.18EE196 pKa = 4.51VAPKK200 pKa = 10.77LIDD203 pKa = 3.5TEE205 pKa = 4.1NTYY208 pKa = 11.11VGLYY212 pKa = 10.17ISDD215 pKa = 3.69TYY217 pKa = 11.1EE218 pKa = 3.88VNDD221 pKa = 4.19RR222 pKa = 11.84LAVTLGGRR230 pKa = 11.84YY231 pKa = 9.39NYY233 pKa = 11.13ASIQLTDD240 pKa = 3.85LTGEE244 pKa = 4.25DD245 pKa = 3.65PALNGTNTYY254 pKa = 10.41EE255 pKa = 4.28RR256 pKa = 11.84FNPAAGLTYY265 pKa = 10.61QLTSTISAYY274 pKa = 10.33GGYY277 pKa = 9.49SEE279 pKa = 5.94ANRR282 pKa = 11.84APVPAEE288 pKa = 3.84LACADD293 pKa = 4.12PEE295 pKa = 4.4RR296 pKa = 11.84PCLIEE301 pKa = 4.31SFLVADD307 pKa = 4.52PPLEE311 pKa = 3.9QVVSNTWEE319 pKa = 3.92LGLRR323 pKa = 11.84GEE325 pKa = 4.62DD326 pKa = 3.3TAADD330 pKa = 4.18GSSRR334 pKa = 11.84WTWNAGLFRR343 pKa = 11.84TYY345 pKa = 10.72TDD347 pKa = 3.85DD348 pKa = 6.22DD349 pKa = 4.68IITVVTNSTRR359 pKa = 11.84GYY361 pKa = 8.42FQNGADD367 pKa = 3.82ILRR370 pKa = 11.84QGIEE374 pKa = 3.9AGLTYY379 pKa = 9.02QTQKK383 pKa = 7.94WQAYY387 pKa = 9.2ANYY390 pKa = 9.91AYY392 pKa = 9.75IDD394 pKa = 3.32ATYY397 pKa = 11.08QNDD400 pKa = 4.99LILNAPDD407 pKa = 3.54NPSARR412 pKa = 11.84EE413 pKa = 4.15SICDD417 pKa = 3.22ASDD420 pKa = 3.58FEE422 pKa = 4.5EE423 pKa = 4.71EE424 pKa = 4.18EE425 pKa = 4.46EE426 pKa = 4.12EE427 pKa = 5.34GEE429 pKa = 4.94DD430 pKa = 3.96EE431 pKa = 6.23GEE433 pKa = 4.11LACIAVRR440 pKa = 11.84PGDD443 pKa = 4.13QLPGIPPHH451 pKa = 6.46RR452 pKa = 11.84FKK454 pKa = 11.36AGMDD458 pKa = 3.24YY459 pKa = 10.71WVTDD463 pKa = 2.81KK464 pKa = 11.07WKK466 pKa = 10.53IGGDD470 pKa = 3.97LIGVSSQYY478 pKa = 10.82FFGDD482 pKa = 3.47EE483 pKa = 4.55ANQQPQLDD491 pKa = 4.74GYY493 pKa = 9.83WRR495 pKa = 11.84VDD497 pKa = 3.03LHH499 pKa = 6.23TSYY502 pKa = 11.05QITPRR507 pKa = 11.84IQLFALANNVFDD519 pKa = 4.29KK520 pKa = 10.79EE521 pKa = 4.29YY522 pKa = 10.89GVFGTFFNLAGANEE536 pKa = 4.2GAEE539 pKa = 4.19ADD541 pKa = 3.99PDD543 pKa = 3.68LGEE546 pKa = 5.77DD547 pKa = 4.16FFTNPRR553 pKa = 11.84TITPAPPAVIYY564 pKa = 10.53GGAKK568 pKa = 8.34IALWW572 pKa = 3.66
Molecular weight: 62.45 kDa
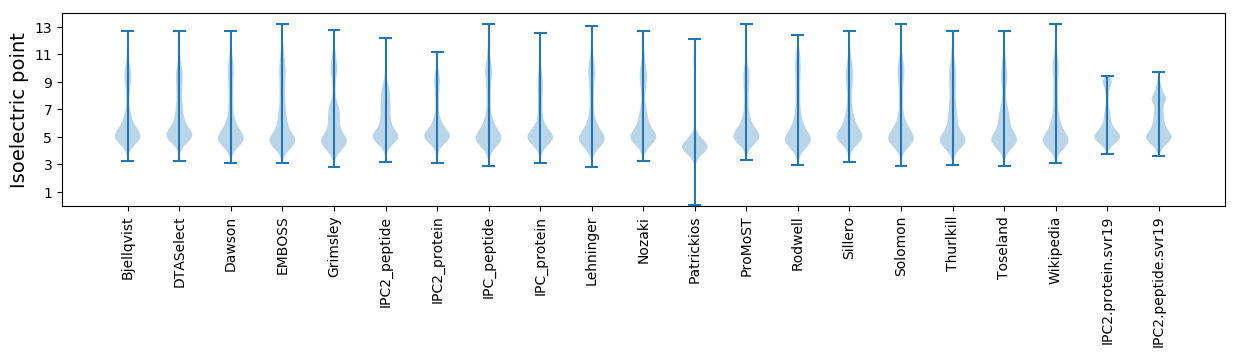
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E2RZN3|A0A1E2RZN3_9RHIZ Uncharacterized protein OS=Methyloligella halotolerans OX=1177755 GN=A7A08_01709 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 9.63IKK4 pKa = 10.84NSIRR8 pKa = 11.84SLRR11 pKa = 11.84KK12 pKa = 7.71RR13 pKa = 11.84HH14 pKa = 5.29RR15 pKa = 11.84ANRR18 pKa = 11.84VVRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84GRR25 pKa = 11.84LYY27 pKa = 10.74VINKK31 pKa = 7.74INRR34 pKa = 11.84RR35 pKa = 11.84FKK37 pKa = 10.84ARR39 pKa = 11.84QGG41 pKa = 3.34
MM1 pKa = 7.66KK2 pKa = 9.63IKK4 pKa = 10.84NSIRR8 pKa = 11.84SLRR11 pKa = 11.84KK12 pKa = 7.71RR13 pKa = 11.84HH14 pKa = 5.29RR15 pKa = 11.84ANRR18 pKa = 11.84VVRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84GRR25 pKa = 11.84LYY27 pKa = 10.74VINKK31 pKa = 7.74INRR34 pKa = 11.84RR35 pKa = 11.84FKK37 pKa = 10.84ARR39 pKa = 11.84QGG41 pKa = 3.34
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
933872 |
29 |
2626 |
294.2 |
32.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.587 ± 0.054 | 0.88 ± 0.015 |
6.036 ± 0.037 | 6.627 ± 0.045 |
3.75 ± 0.029 | 8.629 ± 0.049 |
1.962 ± 0.022 | 5.044 ± 0.031 |
3.818 ± 0.032 | 9.89 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.412 ± 0.023 | 2.64 ± 0.03 |
5.31 ± 0.038 | 3.155 ± 0.025 |
6.646 ± 0.046 | 5.631 ± 0.033 |
5.226 ± 0.035 | 7.01 ± 0.037 |
1.334 ± 0.019 | 2.411 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
