
CRESS virus sp. ctin15
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; unclassified Cressdnaviricota; CRESS viruses
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
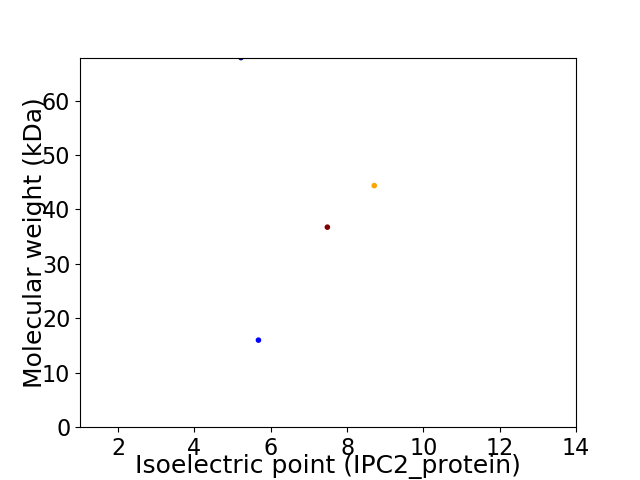
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
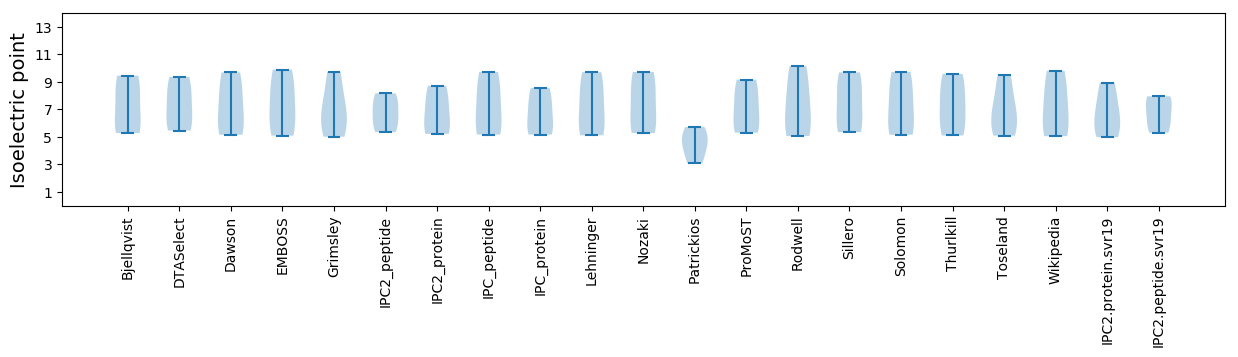
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2WB13|A0A5Q2WB13_9VIRU Rep protein OS=CRESS virus sp. ctin15 OX=2656686 PE=4 SV=1
MM1 pKa = 7.95DD2 pKa = 6.09CYY4 pKa = 11.41SCFYY8 pKa = 9.02TEE10 pKa = 5.16KK11 pKa = 10.13YY12 pKa = 10.21SSSSSDD18 pKa = 3.14SSSDD22 pKa = 3.65DD23 pKa = 4.01SKK25 pKa = 11.82DD26 pKa = 3.29KK27 pKa = 10.97KK28 pKa = 10.48KK29 pKa = 10.82KK30 pKa = 10.3KK31 pKa = 10.29EE32 pKa = 3.94EE33 pKa = 4.01LDD35 pKa = 3.85DD36 pKa = 4.31YY37 pKa = 11.59INFIDD42 pKa = 3.99NPFDD46 pKa = 3.31ISQIDD51 pKa = 3.43IPLSNNVSYY60 pKa = 11.44LDD62 pKa = 3.79YY63 pKa = 10.09PYY65 pKa = 11.22LMPVSSSVSPGYY77 pKa = 9.32TIINRR82 pKa = 11.84DD83 pKa = 3.22IQTLNEE89 pKa = 3.81MRR91 pKa = 11.84PIPFNLNSPFTEE103 pKa = 4.26EE104 pKa = 4.38GLTTALSNYY113 pKa = 9.01FGTGSYY119 pKa = 10.53PIFTKK124 pKa = 10.64ASPSAQYY131 pKa = 10.61LVQEE135 pKa = 4.5GMPIYY140 pKa = 9.92TGPFDD145 pKa = 5.28KK146 pKa = 10.8NGPMNPDD153 pKa = 3.18DD154 pKa = 3.83EE155 pKa = 5.49ARR157 pKa = 11.84FLKK160 pKa = 10.82DD161 pKa = 3.93EE162 pKa = 4.47LEE164 pKa = 4.0PWIRR168 pKa = 11.84LHH170 pKa = 5.97PHH172 pKa = 6.37VIPIIGEE179 pKa = 4.14CVMEE183 pKa = 4.26NPGFHH188 pKa = 7.24PSYY191 pKa = 9.49KK192 pKa = 9.88QRR194 pKa = 11.84GNRR197 pKa = 11.84YY198 pKa = 9.77SFIDD202 pKa = 3.19INGKK206 pKa = 7.58QRR208 pKa = 11.84YY209 pKa = 6.55TYY211 pKa = 10.59GFKK214 pKa = 9.46PTWDD218 pKa = 3.62MFLKK222 pKa = 9.86PDD224 pKa = 4.12KK225 pKa = 10.75YY226 pKa = 10.96PMVVEE231 pKa = 4.15MGNLRR236 pKa = 11.84TSTRR240 pKa = 11.84PVRR243 pKa = 11.84EE244 pKa = 4.02PFRR247 pKa = 11.84GFSPPMYY254 pKa = 10.64ASDD257 pKa = 4.02EE258 pKa = 4.17EE259 pKa = 4.21MDD261 pKa = 4.2GYY263 pKa = 11.01EE264 pKa = 4.8LNPSTLPSVVPPPPPPPPPPPPPTPTPVDD293 pKa = 3.76PNIISPIYY301 pKa = 10.0YY302 pKa = 10.04SYY304 pKa = 11.28PLLFWNEE311 pKa = 3.79PNIISGSSYY320 pKa = 10.41EE321 pKa = 4.04IPQLNSAGLCLNSDD335 pKa = 3.92CSIGGVRR342 pKa = 11.84SIEE345 pKa = 3.98APNIDD350 pKa = 3.68RR351 pKa = 11.84FIIKK355 pKa = 10.07IPDD358 pKa = 3.45NAISRR363 pKa = 11.84VLNVKK368 pKa = 10.0KK369 pKa = 10.79VFYY372 pKa = 9.1KK373 pKa = 10.85TNLNQNLPIIIKK385 pKa = 8.84LKK387 pKa = 9.2CTVLPVVNNYY397 pKa = 9.71PLILTSAPDD406 pKa = 4.36AFPMLYY412 pKa = 9.27TLTIPTVNYY421 pKa = 9.11PLGFTGISILFNTGSSSPSTGGEE444 pKa = 3.79IIFDD448 pKa = 3.72EE449 pKa = 4.71VQTYY453 pKa = 8.34WDD455 pKa = 3.94PRR457 pKa = 11.84VKK459 pKa = 10.86LNLTTHH465 pKa = 6.84IAMNSLFNGGTPNVQADD482 pKa = 3.83NGSATLSGVSKK493 pKa = 9.76TIEE496 pKa = 3.87YY497 pKa = 10.8RR498 pKa = 11.84NGTNNTNLRR507 pKa = 11.84FQRR510 pKa = 11.84NIGLNWNTYY519 pKa = 6.71VQYY522 pKa = 11.3LNRR525 pKa = 11.84TRR527 pKa = 11.84SEE529 pKa = 3.95FNYY532 pKa = 10.42AGYY535 pKa = 11.37AMQGEE540 pKa = 4.7SQYY543 pKa = 11.6SRR545 pKa = 11.84GFTGSANIIGTLGTLGTKK563 pKa = 10.53LNFVAAGVATAAPTTTVTRR582 pKa = 11.84RR583 pKa = 11.84VDD585 pKa = 3.11IFSNDD590 pKa = 3.39YY591 pKa = 11.25LNSQFVNYY599 pKa = 10.25GYY601 pKa = 10.87LHH603 pKa = 6.29YY604 pKa = 10.59IITT607 pKa = 4.05
MM1 pKa = 7.95DD2 pKa = 6.09CYY4 pKa = 11.41SCFYY8 pKa = 9.02TEE10 pKa = 5.16KK11 pKa = 10.13YY12 pKa = 10.21SSSSSDD18 pKa = 3.14SSSDD22 pKa = 3.65DD23 pKa = 4.01SKK25 pKa = 11.82DD26 pKa = 3.29KK27 pKa = 10.97KK28 pKa = 10.48KK29 pKa = 10.82KK30 pKa = 10.3KK31 pKa = 10.29EE32 pKa = 3.94EE33 pKa = 4.01LDD35 pKa = 3.85DD36 pKa = 4.31YY37 pKa = 11.59INFIDD42 pKa = 3.99NPFDD46 pKa = 3.31ISQIDD51 pKa = 3.43IPLSNNVSYY60 pKa = 11.44LDD62 pKa = 3.79YY63 pKa = 10.09PYY65 pKa = 11.22LMPVSSSVSPGYY77 pKa = 9.32TIINRR82 pKa = 11.84DD83 pKa = 3.22IQTLNEE89 pKa = 3.81MRR91 pKa = 11.84PIPFNLNSPFTEE103 pKa = 4.26EE104 pKa = 4.38GLTTALSNYY113 pKa = 9.01FGTGSYY119 pKa = 10.53PIFTKK124 pKa = 10.64ASPSAQYY131 pKa = 10.61LVQEE135 pKa = 4.5GMPIYY140 pKa = 9.92TGPFDD145 pKa = 5.28KK146 pKa = 10.8NGPMNPDD153 pKa = 3.18DD154 pKa = 3.83EE155 pKa = 5.49ARR157 pKa = 11.84FLKK160 pKa = 10.82DD161 pKa = 3.93EE162 pKa = 4.47LEE164 pKa = 4.0PWIRR168 pKa = 11.84LHH170 pKa = 5.97PHH172 pKa = 6.37VIPIIGEE179 pKa = 4.14CVMEE183 pKa = 4.26NPGFHH188 pKa = 7.24PSYY191 pKa = 9.49KK192 pKa = 9.88QRR194 pKa = 11.84GNRR197 pKa = 11.84YY198 pKa = 9.77SFIDD202 pKa = 3.19INGKK206 pKa = 7.58QRR208 pKa = 11.84YY209 pKa = 6.55TYY211 pKa = 10.59GFKK214 pKa = 9.46PTWDD218 pKa = 3.62MFLKK222 pKa = 9.86PDD224 pKa = 4.12KK225 pKa = 10.75YY226 pKa = 10.96PMVVEE231 pKa = 4.15MGNLRR236 pKa = 11.84TSTRR240 pKa = 11.84PVRR243 pKa = 11.84EE244 pKa = 4.02PFRR247 pKa = 11.84GFSPPMYY254 pKa = 10.64ASDD257 pKa = 4.02EE258 pKa = 4.17EE259 pKa = 4.21MDD261 pKa = 4.2GYY263 pKa = 11.01EE264 pKa = 4.8LNPSTLPSVVPPPPPPPPPPPPPTPTPVDD293 pKa = 3.76PNIISPIYY301 pKa = 10.0YY302 pKa = 10.04SYY304 pKa = 11.28PLLFWNEE311 pKa = 3.79PNIISGSSYY320 pKa = 10.41EE321 pKa = 4.04IPQLNSAGLCLNSDD335 pKa = 3.92CSIGGVRR342 pKa = 11.84SIEE345 pKa = 3.98APNIDD350 pKa = 3.68RR351 pKa = 11.84FIIKK355 pKa = 10.07IPDD358 pKa = 3.45NAISRR363 pKa = 11.84VLNVKK368 pKa = 10.0KK369 pKa = 10.79VFYY372 pKa = 9.1KK373 pKa = 10.85TNLNQNLPIIIKK385 pKa = 8.84LKK387 pKa = 9.2CTVLPVVNNYY397 pKa = 9.71PLILTSAPDD406 pKa = 4.36AFPMLYY412 pKa = 9.27TLTIPTVNYY421 pKa = 9.11PLGFTGISILFNTGSSSPSTGGEE444 pKa = 3.79IIFDD448 pKa = 3.72EE449 pKa = 4.71VQTYY453 pKa = 8.34WDD455 pKa = 3.94PRR457 pKa = 11.84VKK459 pKa = 10.86LNLTTHH465 pKa = 6.84IAMNSLFNGGTPNVQADD482 pKa = 3.83NGSATLSGVSKK493 pKa = 9.76TIEE496 pKa = 3.87YY497 pKa = 10.8RR498 pKa = 11.84NGTNNTNLRR507 pKa = 11.84FQRR510 pKa = 11.84NIGLNWNTYY519 pKa = 6.71VQYY522 pKa = 11.3LNRR525 pKa = 11.84TRR527 pKa = 11.84SEE529 pKa = 3.95FNYY532 pKa = 10.42AGYY535 pKa = 11.37AMQGEE540 pKa = 4.7SQYY543 pKa = 11.6SRR545 pKa = 11.84GFTGSANIIGTLGTLGTKK563 pKa = 10.53LNFVAAGVATAAPTTTVTRR582 pKa = 11.84RR583 pKa = 11.84VDD585 pKa = 3.11IFSNDD590 pKa = 3.39YY591 pKa = 11.25LNSQFVNYY599 pKa = 10.25GYY601 pKa = 10.87LHH603 pKa = 6.29YY604 pKa = 10.59IITT607 pKa = 4.05
Molecular weight: 67.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2W8K2|A0A5Q2W8K2_9VIRU Actin OS=CRESS virus sp. ctin15 OX=2656686 PE=4 SV=1
MM1 pKa = 7.52TKK3 pKa = 10.38LSHH6 pKa = 6.83KK7 pKa = 10.5FSLCKK12 pKa = 9.85MISDD16 pKa = 4.5LTTIKK21 pKa = 11.05NPFNTAAFASKK32 pKa = 10.86VPDD35 pKa = 4.02GKK37 pKa = 10.33CGYY40 pKa = 9.95SIGVKK45 pKa = 9.97RR46 pKa = 11.84QVSKK50 pKa = 11.03QIVSSKK56 pKa = 8.86GAKK59 pKa = 9.57IFVLTPTFNCSGFNCDD75 pKa = 3.04VVGNVIDD82 pKa = 4.7GIEE85 pKa = 3.91YY86 pKa = 8.52HH87 pKa = 6.33TFSDD91 pKa = 3.31SDD93 pKa = 3.41ITVRR97 pKa = 11.84APSATALAGTAAATVEE113 pKa = 4.42TNLTLGNNSPEE124 pKa = 4.05KK125 pKa = 9.89WRR127 pKa = 11.84MVSAGMKK134 pKa = 8.56ITLTNNSEE142 pKa = 4.35KK143 pKa = 10.67NDD145 pKa = 3.04GWFEE149 pKa = 4.64AIRR152 pKa = 11.84VNASPSSKK160 pKa = 10.79DD161 pKa = 3.39YY162 pKa = 11.63VFLKK166 pKa = 10.86NVGQDD171 pKa = 3.21EE172 pKa = 5.46GILGVHH178 pKa = 6.89PFFEE182 pKa = 5.03DD183 pKa = 3.65SIIGKK188 pKa = 8.97SEE190 pKa = 3.47SWVNHH195 pKa = 5.48PSYY198 pKa = 10.52VTGKK202 pKa = 10.25LRR204 pKa = 11.84DD205 pKa = 3.56LHH207 pKa = 7.95KK208 pKa = 10.7HH209 pKa = 4.78MFILQQVNGSRR220 pKa = 11.84DD221 pKa = 3.26FCTIRR226 pKa = 11.84QNNTSSGFKK235 pKa = 10.4DD236 pKa = 3.3PQIPIGAFKK245 pKa = 10.62SGPSEE250 pKa = 3.99AFTFKK255 pKa = 10.75NADD258 pKa = 4.24EE259 pKa = 4.43IEE261 pKa = 4.04HH262 pKa = 6.47WADD265 pKa = 3.62DD266 pKa = 3.82SCFDD270 pKa = 3.71VVLVKK275 pKa = 10.55CYY277 pKa = 10.64SATDD281 pKa = 3.73TVGSSTTNLHH291 pKa = 5.37LHH293 pKa = 5.66MVHH296 pKa = 5.2NHH298 pKa = 4.83EE299 pKa = 4.88YY300 pKa = 9.99IYY302 pKa = 11.0DD303 pKa = 3.86SLSTYY308 pKa = 11.22SKK310 pKa = 10.64FQTPTSNMASSVLRR324 pKa = 11.84ATALINSDD332 pKa = 4.29SKK334 pKa = 11.24PSIPFDD340 pKa = 3.63SAPGQLSTCIQACKK354 pKa = 9.14KK355 pKa = 8.92TSNKK359 pKa = 9.89SGRR362 pKa = 11.84SKK364 pKa = 7.6TTRR367 pKa = 11.84SRR369 pKa = 11.84KK370 pKa = 9.13KK371 pKa = 10.83SKK373 pKa = 9.87ICTTKK378 pKa = 9.53RR379 pKa = 11.84RR380 pKa = 11.84SYY382 pKa = 10.98KK383 pKa = 9.58RR384 pKa = 11.84TGCACKK390 pKa = 10.52SPTRR394 pKa = 11.84PPSKK398 pKa = 10.25RR399 pKa = 11.84IRR401 pKa = 11.84KK402 pKa = 9.08VSAA405 pKa = 3.23
MM1 pKa = 7.52TKK3 pKa = 10.38LSHH6 pKa = 6.83KK7 pKa = 10.5FSLCKK12 pKa = 9.85MISDD16 pKa = 4.5LTTIKK21 pKa = 11.05NPFNTAAFASKK32 pKa = 10.86VPDD35 pKa = 4.02GKK37 pKa = 10.33CGYY40 pKa = 9.95SIGVKK45 pKa = 9.97RR46 pKa = 11.84QVSKK50 pKa = 11.03QIVSSKK56 pKa = 8.86GAKK59 pKa = 9.57IFVLTPTFNCSGFNCDD75 pKa = 3.04VVGNVIDD82 pKa = 4.7GIEE85 pKa = 3.91YY86 pKa = 8.52HH87 pKa = 6.33TFSDD91 pKa = 3.31SDD93 pKa = 3.41ITVRR97 pKa = 11.84APSATALAGTAAATVEE113 pKa = 4.42TNLTLGNNSPEE124 pKa = 4.05KK125 pKa = 9.89WRR127 pKa = 11.84MVSAGMKK134 pKa = 8.56ITLTNNSEE142 pKa = 4.35KK143 pKa = 10.67NDD145 pKa = 3.04GWFEE149 pKa = 4.64AIRR152 pKa = 11.84VNASPSSKK160 pKa = 10.79DD161 pKa = 3.39YY162 pKa = 11.63VFLKK166 pKa = 10.86NVGQDD171 pKa = 3.21EE172 pKa = 5.46GILGVHH178 pKa = 6.89PFFEE182 pKa = 5.03DD183 pKa = 3.65SIIGKK188 pKa = 8.97SEE190 pKa = 3.47SWVNHH195 pKa = 5.48PSYY198 pKa = 10.52VTGKK202 pKa = 10.25LRR204 pKa = 11.84DD205 pKa = 3.56LHH207 pKa = 7.95KK208 pKa = 10.7HH209 pKa = 4.78MFILQQVNGSRR220 pKa = 11.84DD221 pKa = 3.26FCTIRR226 pKa = 11.84QNNTSSGFKK235 pKa = 10.4DD236 pKa = 3.3PQIPIGAFKK245 pKa = 10.62SGPSEE250 pKa = 3.99AFTFKK255 pKa = 10.75NADD258 pKa = 4.24EE259 pKa = 4.43IEE261 pKa = 4.04HH262 pKa = 6.47WADD265 pKa = 3.62DD266 pKa = 3.82SCFDD270 pKa = 3.71VVLVKK275 pKa = 10.55CYY277 pKa = 10.64SATDD281 pKa = 3.73TVGSSTTNLHH291 pKa = 5.37LHH293 pKa = 5.66MVHH296 pKa = 5.2NHH298 pKa = 4.83EE299 pKa = 4.88YY300 pKa = 9.99IYY302 pKa = 11.0DD303 pKa = 3.86SLSTYY308 pKa = 11.22SKK310 pKa = 10.64FQTPTSNMASSVLRR324 pKa = 11.84ATALINSDD332 pKa = 4.29SKK334 pKa = 11.24PSIPFDD340 pKa = 3.63SAPGQLSTCIQACKK354 pKa = 9.14KK355 pKa = 8.92TSNKK359 pKa = 9.89SGRR362 pKa = 11.84SKK364 pKa = 7.6TTRR367 pKa = 11.84SRR369 pKa = 11.84KK370 pKa = 9.13KK371 pKa = 10.83SKK373 pKa = 9.87ICTTKK378 pKa = 9.53RR379 pKa = 11.84RR380 pKa = 11.84SYY382 pKa = 10.98KK383 pKa = 9.58RR384 pKa = 11.84TGCACKK390 pKa = 10.52SPTRR394 pKa = 11.84PPSKK398 pKa = 10.25RR399 pKa = 11.84IRR401 pKa = 11.84KK402 pKa = 9.08VSAA405 pKa = 3.23
Molecular weight: 44.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1466 |
136 |
607 |
366.5 |
41.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.775 ± 0.657 | 1.705 ± 0.419 |
5.389 ± 0.238 | 5.116 ± 0.852 |
4.98 ± 0.665 | 5.73 ± 0.506 |
2.115 ± 0.59 | 6.617 ± 0.54 |
6.617 ± 1.228 | 6.821 ± 0.576 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.319 ± 0.215 | 6.617 ± 0.743 |
6.48 ± 1.594 | 2.865 ± 0.357 |
4.638 ± 0.434 | 8.868 ± 1.099 |
7.231 ± 0.849 | 5.935 ± 0.524 |
0.887 ± 0.039 | 4.297 ± 1.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
