
Littorina sp. associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.43
Get precalculated fractions of proteins
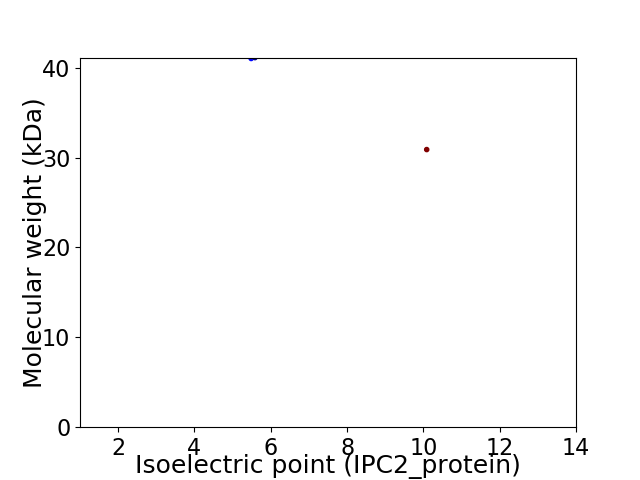
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RLM4|A0A0K1RLM4_9CIRC Putative replication initiation protein OS=Littorina sp. associated circular virus OX=1692253 PE=4 SV=1
MM1 pKa = 7.06PAKK4 pKa = 9.85RR5 pKa = 11.84VYY7 pKa = 9.5QGKK10 pKa = 9.9RR11 pKa = 11.84GKK13 pKa = 9.67RR14 pKa = 11.84WCFTLNNYY22 pKa = 8.26TEE24 pKa = 4.25AEE26 pKa = 4.07KK27 pKa = 11.12DD28 pKa = 3.76VVVAGLIGKK37 pKa = 8.81EE38 pKa = 3.74PVFAKK43 pKa = 10.65VGAEE47 pKa = 4.08VGDD50 pKa = 3.62SGTPHH55 pKa = 6.38LQGFVHH61 pKa = 6.42LKK63 pKa = 9.81EE64 pKa = 4.29RR65 pKa = 11.84EE66 pKa = 4.08SLGGVKK72 pKa = 10.12KK73 pKa = 10.72ILGNRR78 pKa = 11.84CHH80 pKa = 8.77LEE82 pKa = 3.82MARR85 pKa = 11.84GTDD88 pKa = 3.33IEE90 pKa = 4.06NDD92 pKa = 3.47EE93 pKa = 4.38YY94 pKa = 11.52VSKK97 pKa = 11.12GSIVVEE103 pKa = 4.2LGAPVEE109 pKa = 4.38GATEE113 pKa = 4.05KK114 pKa = 11.18GGGDD118 pKa = 3.05TGYY121 pKa = 10.9IMARR125 pKa = 11.84AAAQQLADD133 pKa = 4.58GGDD136 pKa = 3.39LCAIAEE142 pKa = 4.56DD143 pKa = 4.03PEE145 pKa = 4.45LWKK148 pKa = 10.68SYY150 pKa = 8.55CQHH153 pKa = 5.14GRR155 pKa = 11.84AIEE158 pKa = 4.07KK159 pKa = 10.13LAEE162 pKa = 4.03ATRR165 pKa = 11.84KK166 pKa = 9.72SKK168 pKa = 9.55QHH170 pKa = 5.36QLAADD175 pKa = 3.78SLGDD179 pKa = 4.55PILQAWQRR187 pKa = 11.84TLLRR191 pKa = 11.84HH192 pKa = 5.45VLGEE196 pKa = 3.6PDD198 pKa = 3.02MRR200 pKa = 11.84KK201 pKa = 8.68IHH203 pKa = 5.78WMYY206 pKa = 11.42DD207 pKa = 3.17LVGNMGKK214 pKa = 7.63TWMSKK219 pKa = 8.47YY220 pKa = 10.63LCAKK224 pKa = 8.94HH225 pKa = 5.69GAVRR229 pKa = 11.84FEE231 pKa = 4.25NGKK234 pKa = 9.75SADD237 pKa = 2.81IKK239 pKa = 10.1YY240 pKa = 10.05AYY242 pKa = 9.89NGEE245 pKa = 4.22RR246 pKa = 11.84IVIFDD251 pKa = 4.18LSRR254 pKa = 11.84SQEE257 pKa = 3.57DD258 pKa = 3.51HH259 pKa = 6.69FNYY262 pKa = 10.13EE263 pKa = 3.75VLEE266 pKa = 4.2SVKK269 pKa = 10.71NGIMFSPKK277 pKa = 9.25FDD279 pKa = 3.52SKK281 pKa = 10.73MKK283 pKa = 10.23VFAVPHH289 pKa = 5.3VLVFANWGPEE299 pKa = 3.78EE300 pKa = 4.47SKK302 pKa = 10.74LSRR305 pKa = 11.84DD306 pKa = 3.26RR307 pKa = 11.84WDD309 pKa = 3.69VKK311 pKa = 11.18CMTDD315 pKa = 5.27QDD317 pKa = 4.5MEE319 pKa = 4.27WSDD322 pKa = 3.55LASNVIVKK330 pKa = 9.96QEE332 pKa = 3.87GHH334 pKa = 5.93NVLIDD339 pKa = 3.39MTGDD343 pKa = 3.45RR344 pKa = 11.84EE345 pKa = 4.44ADD347 pKa = 3.69TQVMDD352 pKa = 4.06WNSTDD357 pKa = 5.34DD358 pKa = 3.68SDD360 pKa = 5.46CFIVQYY366 pKa = 11.25
MM1 pKa = 7.06PAKK4 pKa = 9.85RR5 pKa = 11.84VYY7 pKa = 9.5QGKK10 pKa = 9.9RR11 pKa = 11.84GKK13 pKa = 9.67RR14 pKa = 11.84WCFTLNNYY22 pKa = 8.26TEE24 pKa = 4.25AEE26 pKa = 4.07KK27 pKa = 11.12DD28 pKa = 3.76VVVAGLIGKK37 pKa = 8.81EE38 pKa = 3.74PVFAKK43 pKa = 10.65VGAEE47 pKa = 4.08VGDD50 pKa = 3.62SGTPHH55 pKa = 6.38LQGFVHH61 pKa = 6.42LKK63 pKa = 9.81EE64 pKa = 4.29RR65 pKa = 11.84EE66 pKa = 4.08SLGGVKK72 pKa = 10.12KK73 pKa = 10.72ILGNRR78 pKa = 11.84CHH80 pKa = 8.77LEE82 pKa = 3.82MARR85 pKa = 11.84GTDD88 pKa = 3.33IEE90 pKa = 4.06NDD92 pKa = 3.47EE93 pKa = 4.38YY94 pKa = 11.52VSKK97 pKa = 11.12GSIVVEE103 pKa = 4.2LGAPVEE109 pKa = 4.38GATEE113 pKa = 4.05KK114 pKa = 11.18GGGDD118 pKa = 3.05TGYY121 pKa = 10.9IMARR125 pKa = 11.84AAAQQLADD133 pKa = 4.58GGDD136 pKa = 3.39LCAIAEE142 pKa = 4.56DD143 pKa = 4.03PEE145 pKa = 4.45LWKK148 pKa = 10.68SYY150 pKa = 8.55CQHH153 pKa = 5.14GRR155 pKa = 11.84AIEE158 pKa = 4.07KK159 pKa = 10.13LAEE162 pKa = 4.03ATRR165 pKa = 11.84KK166 pKa = 9.72SKK168 pKa = 9.55QHH170 pKa = 5.36QLAADD175 pKa = 3.78SLGDD179 pKa = 4.55PILQAWQRR187 pKa = 11.84TLLRR191 pKa = 11.84HH192 pKa = 5.45VLGEE196 pKa = 3.6PDD198 pKa = 3.02MRR200 pKa = 11.84KK201 pKa = 8.68IHH203 pKa = 5.78WMYY206 pKa = 11.42DD207 pKa = 3.17LVGNMGKK214 pKa = 7.63TWMSKK219 pKa = 8.47YY220 pKa = 10.63LCAKK224 pKa = 8.94HH225 pKa = 5.69GAVRR229 pKa = 11.84FEE231 pKa = 4.25NGKK234 pKa = 9.75SADD237 pKa = 2.81IKK239 pKa = 10.1YY240 pKa = 10.05AYY242 pKa = 9.89NGEE245 pKa = 4.22RR246 pKa = 11.84IVIFDD251 pKa = 4.18LSRR254 pKa = 11.84SQEE257 pKa = 3.57DD258 pKa = 3.51HH259 pKa = 6.69FNYY262 pKa = 10.13EE263 pKa = 3.75VLEE266 pKa = 4.2SVKK269 pKa = 10.71NGIMFSPKK277 pKa = 9.25FDD279 pKa = 3.52SKK281 pKa = 10.73MKK283 pKa = 10.23VFAVPHH289 pKa = 5.3VLVFANWGPEE299 pKa = 3.78EE300 pKa = 4.47SKK302 pKa = 10.74LSRR305 pKa = 11.84DD306 pKa = 3.26RR307 pKa = 11.84WDD309 pKa = 3.69VKK311 pKa = 11.18CMTDD315 pKa = 5.27QDD317 pKa = 4.5MEE319 pKa = 4.27WSDD322 pKa = 3.55LASNVIVKK330 pKa = 9.96QEE332 pKa = 3.87GHH334 pKa = 5.93NVLIDD339 pKa = 3.39MTGDD343 pKa = 3.45RR344 pKa = 11.84EE345 pKa = 4.44ADD347 pKa = 3.69TQVMDD352 pKa = 4.06WNSTDD357 pKa = 5.34DD358 pKa = 3.68SDD360 pKa = 5.46CFIVQYY366 pKa = 11.25
Molecular weight: 41.03 kDa
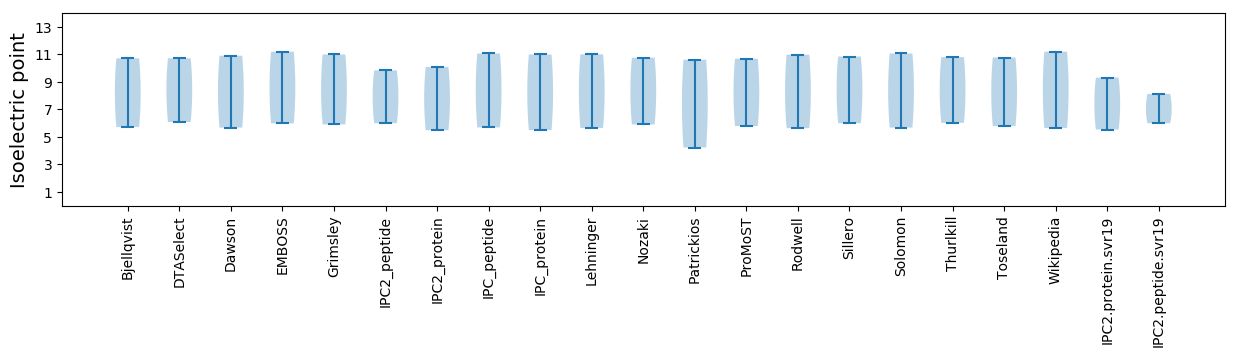
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RLM4|A0A0K1RLM4_9CIRC Putative replication initiation protein OS=Littorina sp. associated circular virus OX=1692253 PE=4 SV=1
MM1 pKa = 8.11AKK3 pKa = 10.23RR4 pKa = 11.84KK5 pKa = 5.39MTSYY9 pKa = 10.53FKK11 pKa = 10.5SKK13 pKa = 10.08RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84FTAPSRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84GPFGGGNKK33 pKa = 7.94TSRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84FTRR42 pKa = 11.84TRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84TRR50 pKa = 11.84NQPSRR55 pKa = 11.84YY56 pKa = 7.72QVFNARR62 pKa = 11.84IYY64 pKa = 8.68YY65 pKa = 8.57TVAADD70 pKa = 5.08ANGQLYY76 pKa = 9.76RR77 pKa = 11.84NNYY80 pKa = 9.39LSVHH84 pKa = 5.91GAIQEE89 pKa = 4.01IQNNNRR95 pKa = 11.84AIYY98 pKa = 10.47DD99 pKa = 3.78SVLTNWDD106 pKa = 3.17KK107 pKa = 11.4FKK109 pKa = 10.94FLSKK113 pKa = 9.23TEE115 pKa = 4.42RR116 pKa = 11.84IFMSDD121 pKa = 2.89TSPYY125 pKa = 10.66NDD127 pKa = 3.55TARR130 pKa = 11.84DD131 pKa = 3.5RR132 pKa = 11.84ATTLYY137 pKa = 10.29HH138 pKa = 6.95VYY140 pKa = 10.73DD141 pKa = 4.46PDD143 pKa = 3.55SRR145 pKa = 11.84GRR147 pKa = 11.84RR148 pKa = 11.84MAPSEE153 pKa = 4.15MMKK156 pKa = 9.75MPACHH161 pKa = 6.39WKK163 pKa = 10.34FIRR166 pKa = 11.84PYY168 pKa = 9.94QVYY171 pKa = 10.04KK172 pKa = 9.85STLRR176 pKa = 11.84PVFQIPAAGAVLDD189 pKa = 4.32GGEE192 pKa = 4.14ISTGIPRR199 pKa = 11.84VDD201 pKa = 3.93NPWRR205 pKa = 11.84DD206 pKa = 3.61CANFTPTGLASLEE219 pKa = 4.22SHH221 pKa = 6.4NGAMYY226 pKa = 10.46SISTSPNQQFTVIRR240 pKa = 11.84TYY242 pKa = 10.71KK243 pKa = 10.2IAVTTPRR250 pKa = 11.84YY251 pKa = 8.1GQLYY255 pKa = 9.52QGQDD259 pKa = 2.79NTNYY263 pKa = 9.99IEE265 pKa = 3.94
MM1 pKa = 8.11AKK3 pKa = 10.23RR4 pKa = 11.84KK5 pKa = 5.39MTSYY9 pKa = 10.53FKK11 pKa = 10.5SKK13 pKa = 10.08RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84FTAPSRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84GPFGGGNKK33 pKa = 7.94TSRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84FTRR42 pKa = 11.84TRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84TRR50 pKa = 11.84NQPSRR55 pKa = 11.84YY56 pKa = 7.72QVFNARR62 pKa = 11.84IYY64 pKa = 8.68YY65 pKa = 8.57TVAADD70 pKa = 5.08ANGQLYY76 pKa = 9.76RR77 pKa = 11.84NNYY80 pKa = 9.39LSVHH84 pKa = 5.91GAIQEE89 pKa = 4.01IQNNNRR95 pKa = 11.84AIYY98 pKa = 10.47DD99 pKa = 3.78SVLTNWDD106 pKa = 3.17KK107 pKa = 11.4FKK109 pKa = 10.94FLSKK113 pKa = 9.23TEE115 pKa = 4.42RR116 pKa = 11.84IFMSDD121 pKa = 2.89TSPYY125 pKa = 10.66NDD127 pKa = 3.55TARR130 pKa = 11.84DD131 pKa = 3.5RR132 pKa = 11.84ATTLYY137 pKa = 10.29HH138 pKa = 6.95VYY140 pKa = 10.73DD141 pKa = 4.46PDD143 pKa = 3.55SRR145 pKa = 11.84GRR147 pKa = 11.84RR148 pKa = 11.84MAPSEE153 pKa = 4.15MMKK156 pKa = 9.75MPACHH161 pKa = 6.39WKK163 pKa = 10.34FIRR166 pKa = 11.84PYY168 pKa = 9.94QVYY171 pKa = 10.04KK172 pKa = 9.85STLRR176 pKa = 11.84PVFQIPAAGAVLDD189 pKa = 4.32GGEE192 pKa = 4.14ISTGIPRR199 pKa = 11.84VDD201 pKa = 3.93NPWRR205 pKa = 11.84DD206 pKa = 3.61CANFTPTGLASLEE219 pKa = 4.22SHH221 pKa = 6.4NGAMYY226 pKa = 10.46SISTSPNQQFTVIRR240 pKa = 11.84TYY242 pKa = 10.71KK243 pKa = 10.2IAVTTPRR250 pKa = 11.84YY251 pKa = 8.1GQLYY255 pKa = 9.52QGQDD259 pKa = 2.79NTNYY263 pKa = 9.99IEE265 pKa = 3.94
Molecular weight: 30.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
631 |
265 |
366 |
315.5 |
35.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.607 ± 0.27 | 1.426 ± 0.415 |
6.339 ± 1.12 | 5.23 ± 1.834 |
3.645 ± 0.546 | 7.607 ± 1.204 |
2.377 ± 0.537 | 4.596 ± 0.192 |
6.339 ± 1.12 | 5.705 ± 1.195 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.328 ± 0.191 | 4.754 ± 1.027 |
3.962 ± 1.05 | 4.12 ± 0.252 |
8.399 ± 2.974 | 6.181 ± 0.612 |
5.705 ± 1.839 | 6.339 ± 1.353 |
1.902 ± 0.476 | 4.437 ± 1.223 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
