
Triticum urartu (Red wild einkorn) (Crithodium urartu)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Triticodae; Triticeae; Triticinae; Triticum
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
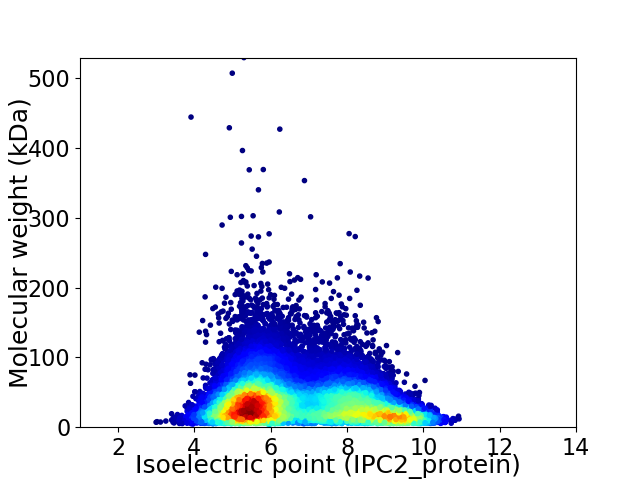
Virtual 2D-PAGE plot for 33312 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
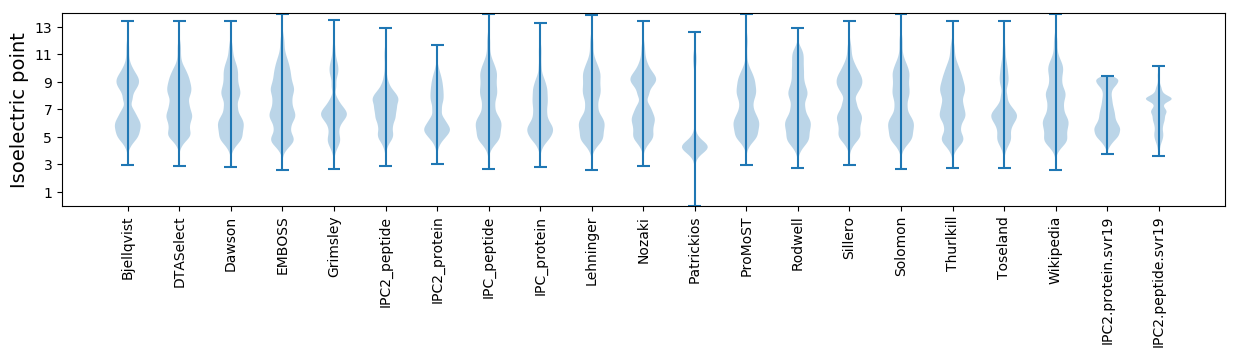
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M8AIS2|M8AIS2_TRIUA Cucumisin OS=Triticum urartu OX=4572 GN=TRIUR3_23719 PE=3 SV=1
MM1 pKa = 7.76APTNSPPIHH10 pKa = 6.27SAQGGAVHH18 pKa = 6.85GVGNDD23 pKa = 3.34IVSHH27 pKa = 6.52ASASLCTAADD37 pKa = 3.7EE38 pKa = 4.88ASVFGQGPVDD48 pKa = 3.4RR49 pKa = 11.84AVIEE53 pKa = 4.16MVEE56 pKa = 4.14VAEE59 pKa = 4.28NAVVLPQIHH68 pKa = 6.09NAVEE72 pKa = 4.11EE73 pKa = 4.23TGEE76 pKa = 4.62FDD78 pKa = 4.04PLDD81 pKa = 3.9YY82 pKa = 10.9NAPAGANEE90 pKa = 4.5LADD93 pKa = 4.18GDD95 pKa = 4.32NEE97 pKa = 4.79SGTGNVDD104 pKa = 3.29RR105 pKa = 11.84AAADD109 pKa = 4.15DD110 pKa = 5.28DD111 pKa = 4.56VDD113 pKa = 4.41SPPSKK118 pKa = 10.84PEE120 pKa = 3.64DD121 pKa = 3.29LVYY124 pKa = 10.72LFHH127 pKa = 7.15GSYY130 pKa = 10.87GVACVV135 pKa = 3.32
MM1 pKa = 7.76APTNSPPIHH10 pKa = 6.27SAQGGAVHH18 pKa = 6.85GVGNDD23 pKa = 3.34IVSHH27 pKa = 6.52ASASLCTAADD37 pKa = 3.7EE38 pKa = 4.88ASVFGQGPVDD48 pKa = 3.4RR49 pKa = 11.84AVIEE53 pKa = 4.16MVEE56 pKa = 4.14VAEE59 pKa = 4.28NAVVLPQIHH68 pKa = 6.09NAVEE72 pKa = 4.11EE73 pKa = 4.23TGEE76 pKa = 4.62FDD78 pKa = 4.04PLDD81 pKa = 3.9YY82 pKa = 10.9NAPAGANEE90 pKa = 4.5LADD93 pKa = 4.18GDD95 pKa = 4.32NEE97 pKa = 4.79SGTGNVDD104 pKa = 3.29RR105 pKa = 11.84AAADD109 pKa = 4.15DD110 pKa = 5.28DD111 pKa = 4.56VDD113 pKa = 4.41SPPSKK118 pKa = 10.84PEE120 pKa = 3.64DD121 pKa = 3.29LVYY124 pKa = 10.72LFHH127 pKa = 7.15GSYY130 pKa = 10.87GVACVV135 pKa = 3.32
Molecular weight: 13.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T1LP83|T1LP83_TRIUA Uncharacterized protein OS=Triticum urartu OX=4572 PE=4 SV=1
TT1 pKa = 7.5AFTPRR6 pKa = 11.84ATLVSLVVTRR16 pKa = 11.84TRR18 pKa = 11.84LPRR21 pKa = 11.84ATSLRR26 pKa = 11.84GVSSTSPHH34 pKa = 6.55ASWLHH39 pKa = 5.06TSSITRR45 pKa = 11.84NIFFPVKK52 pKa = 9.36ASLRR56 pKa = 11.84IGQSTANTFSLRR68 pKa = 11.84CFPPARR74 pKa = 11.84VMAVLRR80 pKa = 11.84SSTLSKK86 pKa = 10.64SWLTLNHH93 pKa = 6.36IFLSKK98 pKa = 10.18LAFMASSLKK107 pKa = 9.28TSRR110 pKa = 11.84ARR112 pKa = 11.84VVLPIPPTPTMAKK125 pKa = 9.63TMTLLLSALLVSASGLLTMISTSALVSSSIFSPP158 pKa = 3.74
TT1 pKa = 7.5AFTPRR6 pKa = 11.84ATLVSLVVTRR16 pKa = 11.84TRR18 pKa = 11.84LPRR21 pKa = 11.84ATSLRR26 pKa = 11.84GVSSTSPHH34 pKa = 6.55ASWLHH39 pKa = 5.06TSSITRR45 pKa = 11.84NIFFPVKK52 pKa = 9.36ASLRR56 pKa = 11.84IGQSTANTFSLRR68 pKa = 11.84CFPPARR74 pKa = 11.84VMAVLRR80 pKa = 11.84SSTLSKK86 pKa = 10.64SWLTLNHH93 pKa = 6.36IFLSKK98 pKa = 10.18LAFMASSLKK107 pKa = 9.28TSRR110 pKa = 11.84ARR112 pKa = 11.84VVLPIPPTPTMAKK125 pKa = 9.63TMTLLLSALLVSASGLLTMISTSALVSSSIFSPP158 pKa = 3.74
Molecular weight: 16.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12117229 |
29 |
5024 |
363.7 |
40.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.321 ± 0.015 | 2.016 ± 0.007 |
5.482 ± 0.012 | 6.11 ± 0.014 |
3.789 ± 0.008 | 7.218 ± 0.013 |
2.562 ± 0.006 | 4.658 ± 0.01 |
5.183 ± 0.014 | 9.791 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.563 ± 0.005 | 3.631 ± 0.009 |
5.209 ± 0.015 | 3.547 ± 0.011 |
5.996 ± 0.012 | 8.053 ± 0.014 |
4.932 ± 0.009 | 6.908 ± 0.009 |
1.355 ± 0.005 | 2.673 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
