
Beihai weivirus-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
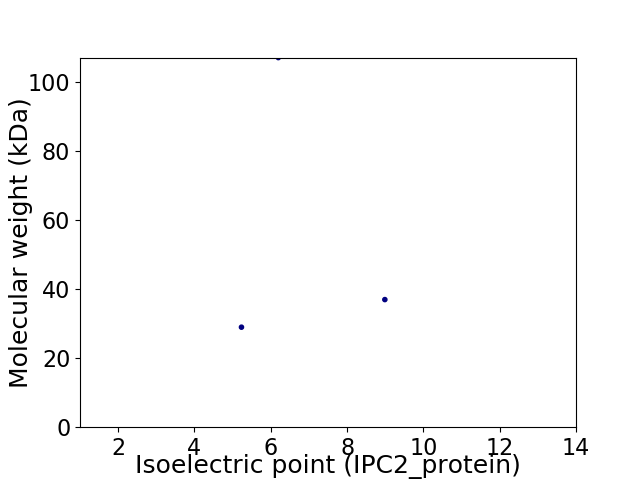
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
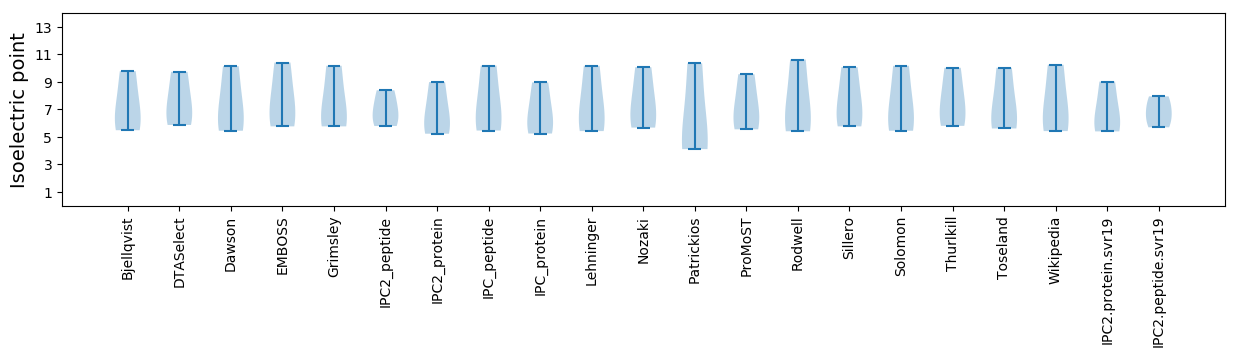
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KL03|A0A1L3KL03_9VIRU RNA-dependent RNA polymerase OS=Beihai weivirus-like virus 2 OX=1922748 PE=4 SV=1
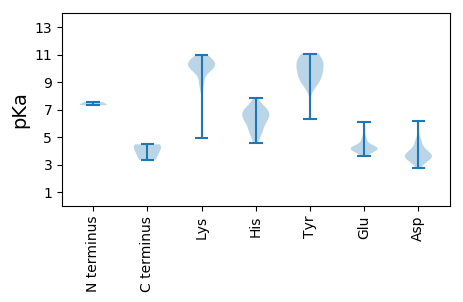
MM1 pKa = 7.31LQGDD5 pKa = 4.29VANTPALALVGTQPPPGLSTCSVRR29 pKa = 11.84QTGVLGGVHH38 pKa = 6.63ARR40 pKa = 11.84TAGRR44 pKa = 11.84NIPGPVPGSPAWLVWVHH61 pKa = 6.38DD62 pKa = 4.7HH63 pKa = 7.18FDD65 pKa = 3.71HH66 pKa = 7.85PDD68 pKa = 3.5GEE70 pKa = 5.1GVRR73 pKa = 11.84EE74 pKa = 3.76THH76 pKa = 6.29GRR78 pKa = 11.84GEE80 pKa = 4.15AARR83 pKa = 11.84HH84 pKa = 5.45GEE86 pKa = 4.34GEE88 pKa = 4.34GSSVAGSSQHH98 pKa = 6.38RR99 pKa = 11.84GQRR102 pKa = 11.84TLEE105 pKa = 3.85DD106 pKa = 3.17RR107 pKa = 11.84EE108 pKa = 4.03YY109 pKa = 9.37RR110 pKa = 11.84TCGTQVFGTGHH121 pKa = 7.09DD122 pKa = 3.63NPRR125 pKa = 11.84GQGVEE130 pKa = 4.12HH131 pKa = 6.81NGHH134 pKa = 6.22HH135 pKa = 6.42RR136 pKa = 11.84RR137 pKa = 11.84PSFIAYY143 pKa = 9.72DD144 pKa = 3.54IVVHH148 pKa = 6.88DD149 pKa = 4.9FGIPPARR156 pKa = 11.84VGSLADD162 pKa = 3.3GHH164 pKa = 5.62VAPTVGEE171 pKa = 4.07QTILEE176 pKa = 4.39AASDD180 pKa = 3.76AGDD183 pKa = 3.33VDD185 pKa = 3.95GEE187 pKa = 4.39RR188 pKa = 11.84VQGGARR194 pKa = 11.84EE195 pKa = 3.99AAVGGGQPWEE205 pKa = 4.1AGLRR209 pKa = 11.84LARR212 pKa = 11.84GRR214 pKa = 11.84LGGQTDD220 pKa = 3.93DD221 pKa = 4.35EE222 pKa = 6.1DD223 pKa = 5.43DD224 pKa = 4.27GFVGARR230 pKa = 11.84WMGCMVWSSGVHH242 pKa = 4.95GHH244 pKa = 7.34DD245 pKa = 4.66EE246 pKa = 4.52VPCDD250 pKa = 3.83DD251 pKa = 5.1LLSEE255 pKa = 4.26THH257 pKa = 6.56ASARR261 pKa = 11.84LLEE264 pKa = 4.44RR265 pKa = 11.84TDD267 pKa = 3.51SQFHH271 pKa = 6.22FGWW274 pKa = 4.47
MM1 pKa = 7.31LQGDD5 pKa = 4.29VANTPALALVGTQPPPGLSTCSVRR29 pKa = 11.84QTGVLGGVHH38 pKa = 6.63ARR40 pKa = 11.84TAGRR44 pKa = 11.84NIPGPVPGSPAWLVWVHH61 pKa = 6.38DD62 pKa = 4.7HH63 pKa = 7.18FDD65 pKa = 3.71HH66 pKa = 7.85PDD68 pKa = 3.5GEE70 pKa = 5.1GVRR73 pKa = 11.84EE74 pKa = 3.76THH76 pKa = 6.29GRR78 pKa = 11.84GEE80 pKa = 4.15AARR83 pKa = 11.84HH84 pKa = 5.45GEE86 pKa = 4.34GEE88 pKa = 4.34GSSVAGSSQHH98 pKa = 6.38RR99 pKa = 11.84GQRR102 pKa = 11.84TLEE105 pKa = 3.85DD106 pKa = 3.17RR107 pKa = 11.84EE108 pKa = 4.03YY109 pKa = 9.37RR110 pKa = 11.84TCGTQVFGTGHH121 pKa = 7.09DD122 pKa = 3.63NPRR125 pKa = 11.84GQGVEE130 pKa = 4.12HH131 pKa = 6.81NGHH134 pKa = 6.22HH135 pKa = 6.42RR136 pKa = 11.84RR137 pKa = 11.84PSFIAYY143 pKa = 9.72DD144 pKa = 3.54IVVHH148 pKa = 6.88DD149 pKa = 4.9FGIPPARR156 pKa = 11.84VGSLADD162 pKa = 3.3GHH164 pKa = 5.62VAPTVGEE171 pKa = 4.07QTILEE176 pKa = 4.39AASDD180 pKa = 3.76AGDD183 pKa = 3.33VDD185 pKa = 3.95GEE187 pKa = 4.39RR188 pKa = 11.84VQGGARR194 pKa = 11.84EE195 pKa = 3.99AAVGGGQPWEE205 pKa = 4.1AGLRR209 pKa = 11.84LARR212 pKa = 11.84GRR214 pKa = 11.84LGGQTDD220 pKa = 3.93DD221 pKa = 4.35EE222 pKa = 6.1DD223 pKa = 5.43DD224 pKa = 4.27GFVGARR230 pKa = 11.84WMGCMVWSSGVHH242 pKa = 4.95GHH244 pKa = 7.34DD245 pKa = 4.66EE246 pKa = 4.52VPCDD250 pKa = 3.83DD251 pKa = 5.1LLSEE255 pKa = 4.26THH257 pKa = 6.56ASARR261 pKa = 11.84LLEE264 pKa = 4.44RR265 pKa = 11.84TDD267 pKa = 3.51SQFHH271 pKa = 6.22FGWW274 pKa = 4.47
Molecular weight: 28.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KL47|A0A1L3KL47_9VIRU Putative capsid protein OS=Beihai weivirus-like virus 2 OX=1922748 PE=4 SV=1
MM1 pKa = 7.55AKK3 pKa = 10.18KK4 pKa = 9.76FVKK7 pKa = 10.26KK8 pKa = 10.4LPQRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PVIKK19 pKa = 10.17RR20 pKa = 11.84NMKK23 pKa = 8.84ATRR26 pKa = 11.84SHH28 pKa = 6.05ATRR31 pKa = 11.84VLAQGAGAITKK42 pKa = 9.97KK43 pKa = 10.85AFGAGKK49 pKa = 8.52KK50 pKa = 4.91TTTWRR55 pKa = 11.84MALRR59 pKa = 11.84RR60 pKa = 11.84GLNARR65 pKa = 11.84LPYY68 pKa = 10.31HH69 pKa = 6.99LGLPRR74 pKa = 11.84PVGPYY79 pKa = 9.52QVIRR83 pKa = 11.84TTKK86 pKa = 9.98LHH88 pKa = 5.24STSARR93 pKa = 11.84VVVFCPLMQKK103 pKa = 9.86WNGQDD108 pKa = 3.43DD109 pKa = 3.85APRR112 pKa = 11.84WFEE115 pKa = 3.66ACGIEE120 pKa = 5.86DD121 pKa = 3.57VDD123 pKa = 3.66ATKK126 pKa = 10.41PINDD130 pKa = 3.58ATGNAHH136 pKa = 6.87MIGMPLTGIGSAAEE150 pKa = 4.03VVPAAMTVQVMCSDD164 pKa = 4.8PLQTAQGIFTMGRR177 pKa = 11.84VSQQLPLGGEE187 pKa = 4.05TRR189 pKa = 11.84SWDD192 pKa = 3.55TFKK195 pKa = 10.32TQFDD199 pKa = 4.36AYY201 pKa = 10.05FKK203 pKa = 10.74PRR205 pKa = 11.84LLTGGKK211 pKa = 9.25LALRR215 pKa = 11.84GVTCNAIPLNMNEE228 pKa = 4.03FSEE231 pKa = 4.8FASPQAAEE239 pKa = 5.38NNFIWNDD246 pKa = 3.7TVVPAALSPIVFTKK260 pKa = 10.83DD261 pKa = 2.88PGEE264 pKa = 4.29VGSSITFLVTIEE276 pKa = 3.6WRR278 pKa = 11.84VRR280 pKa = 11.84FDD282 pKa = 3.6PFHH285 pKa = 6.87PAAASHH291 pKa = 6.21TFHH294 pKa = 7.55PSTPDD299 pKa = 3.36SLWNRR304 pKa = 11.84VIGAASSMGHH314 pKa = 5.18GVEE317 pKa = 5.65DD318 pKa = 3.61IADD321 pKa = 3.78EE322 pKa = 4.24VAEE325 pKa = 4.62LGADD329 pKa = 3.34AAAAAVVGALAFF341 pKa = 4.18
MM1 pKa = 7.55AKK3 pKa = 10.18KK4 pKa = 9.76FVKK7 pKa = 10.26KK8 pKa = 10.4LPQRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PVIKK19 pKa = 10.17RR20 pKa = 11.84NMKK23 pKa = 8.84ATRR26 pKa = 11.84SHH28 pKa = 6.05ATRR31 pKa = 11.84VLAQGAGAITKK42 pKa = 9.97KK43 pKa = 10.85AFGAGKK49 pKa = 8.52KK50 pKa = 4.91TTTWRR55 pKa = 11.84MALRR59 pKa = 11.84RR60 pKa = 11.84GLNARR65 pKa = 11.84LPYY68 pKa = 10.31HH69 pKa = 6.99LGLPRR74 pKa = 11.84PVGPYY79 pKa = 9.52QVIRR83 pKa = 11.84TTKK86 pKa = 9.98LHH88 pKa = 5.24STSARR93 pKa = 11.84VVVFCPLMQKK103 pKa = 9.86WNGQDD108 pKa = 3.43DD109 pKa = 3.85APRR112 pKa = 11.84WFEE115 pKa = 3.66ACGIEE120 pKa = 5.86DD121 pKa = 3.57VDD123 pKa = 3.66ATKK126 pKa = 10.41PINDD130 pKa = 3.58ATGNAHH136 pKa = 6.87MIGMPLTGIGSAAEE150 pKa = 4.03VVPAAMTVQVMCSDD164 pKa = 4.8PLQTAQGIFTMGRR177 pKa = 11.84VSQQLPLGGEE187 pKa = 4.05TRR189 pKa = 11.84SWDD192 pKa = 3.55TFKK195 pKa = 10.32TQFDD199 pKa = 4.36AYY201 pKa = 10.05FKK203 pKa = 10.74PRR205 pKa = 11.84LLTGGKK211 pKa = 9.25LALRR215 pKa = 11.84GVTCNAIPLNMNEE228 pKa = 4.03FSEE231 pKa = 4.8FASPQAAEE239 pKa = 5.38NNFIWNDD246 pKa = 3.7TVVPAALSPIVFTKK260 pKa = 10.83DD261 pKa = 2.88PGEE264 pKa = 4.29VGSSITFLVTIEE276 pKa = 3.6WRR278 pKa = 11.84VRR280 pKa = 11.84FDD282 pKa = 3.6PFHH285 pKa = 6.87PAAASHH291 pKa = 6.21TFHH294 pKa = 7.55PSTPDD299 pKa = 3.36SLWNRR304 pKa = 11.84VIGAASSMGHH314 pKa = 5.18GVEE317 pKa = 5.65DD318 pKa = 3.61IADD321 pKa = 3.78EE322 pKa = 4.24VAEE325 pKa = 4.62LGADD329 pKa = 3.34AAAAAVVGALAFF341 pKa = 4.18
Molecular weight: 36.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1576 |
274 |
961 |
525.3 |
57.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.025 ± 0.988 | 1.396 ± 0.077 |
5.393 ± 0.595 | 6.853 ± 1.332 |
3.871 ± 0.472 | 9.581 ± 2.098 |
2.728 ± 1.099 | 3.68 ± 0.566 |
4.759 ± 1.537 | 7.551 ± 0.731 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.728 ± 0.498 | 2.538 ± 0.401 |
5.901 ± 0.306 | 3.68 ± 0.215 |
7.043 ± 0.316 | 5.33 ± 0.118 |
6.028 ± 0.55 | 7.234 ± 0.573 |
2.284 ± 0.102 | 1.396 ± 0.341 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
