
Pseudomonas phage pf8_ST274-AUS411
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; unclassified Inoviridae
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
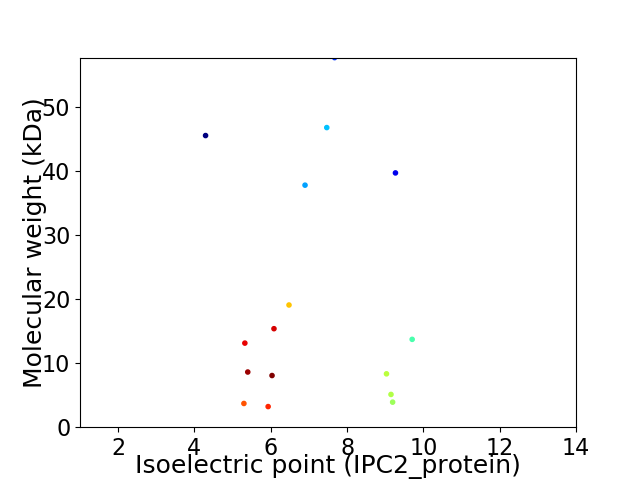
Virtual 2D-PAGE plot for 16 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B9J4B2|A0A6B9J4B2_9VIRU Uncharacterized protein OS=Pseudomonas phage pf8_ST274-AUS411 OX=2686285 PE=4 SV=1
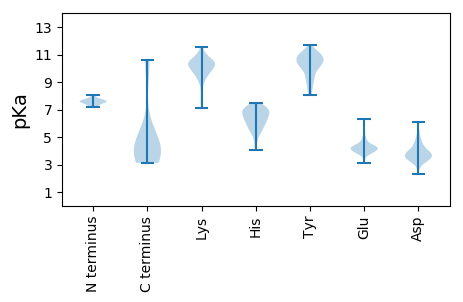
MM1 pKa = 7.79RR2 pKa = 11.84IKK4 pKa = 10.19RR5 pKa = 11.84TLLVLLTLFMSVCASAEE22 pKa = 3.87DD23 pKa = 4.78YY24 pKa = 10.1YY25 pKa = 10.62WPRR28 pKa = 11.84GTQKK32 pKa = 9.82YY33 pKa = 8.53GSYY36 pKa = 9.51MEE38 pKa = 4.29VVEE41 pKa = 4.74EE42 pKa = 3.89ARR44 pKa = 11.84KK45 pKa = 9.19AAIANNPGYY54 pKa = 10.76SRR56 pKa = 11.84VEE58 pKa = 4.01AVRR61 pKa = 11.84VIYY64 pKa = 8.8PANGRR69 pKa = 11.84QDD71 pKa = 3.05MATYY75 pKa = 10.55GLEE78 pKa = 4.63FYY80 pKa = 10.71CLSQGVEE87 pKa = 4.42RR88 pKa = 11.84MCSTSYY94 pKa = 11.27NNPVYY99 pKa = 10.49RR100 pKa = 11.84KK101 pKa = 10.45GEE103 pKa = 3.98GCTAPKK109 pKa = 10.31IPDD112 pKa = 3.7EE113 pKa = 4.32TTGTCKK119 pKa = 10.35EE120 pKa = 4.38PPTPPEE126 pKa = 4.1DD127 pKa = 4.36CIKK130 pKa = 11.06GLTDD134 pKa = 4.65LFSSPPSNIFVSGGRR149 pKa = 11.84NFVNSSPPTGCKK161 pKa = 9.91NGCQYY166 pKa = 11.41LPTTSKK172 pKa = 7.66TTSCYY177 pKa = 10.13RR178 pKa = 11.84YY179 pKa = 9.48PGSDD183 pKa = 2.89NQGFCNYY190 pKa = 9.86VLMTDD195 pKa = 4.32GSACAADD202 pKa = 3.63SGNPGMTGPSLNDD215 pKa = 3.57TPPTNPDD222 pKa = 3.95EE223 pKa = 4.85PPSDD227 pKa = 4.41PNDD230 pKa = 4.17PGCPPGYY237 pKa = 8.96SWSGTTCVKK246 pKa = 10.27TPTDD250 pKa = 3.57PTEE253 pKa = 4.25PGGDD257 pKa = 3.65GGDD260 pKa = 3.91GGDD263 pKa = 4.12GGNTGGGDD271 pKa = 3.29GGGDD275 pKa = 2.96NGGGNDD281 pKa = 3.77NGGGDD286 pKa = 3.94GGTGGSDD293 pKa = 2.98GSGGNGEE300 pKa = 4.28GGGDD304 pKa = 3.57GSGGGDD310 pKa = 3.27GSGGGTGGGDD320 pKa = 3.45GGDD323 pKa = 3.81GGDD326 pKa = 5.45GGNCDD331 pKa = 4.17PAKK334 pKa = 10.16QGCSPGPAGPGGEE347 pKa = 4.21LKK349 pKa = 10.5EE350 pKa = 4.48PKK352 pKa = 9.97PGTWDD357 pKa = 3.36DD358 pKa = 6.11AIATWEE364 pKa = 4.22QKK366 pKa = 10.35VEE368 pKa = 4.01QAKK371 pKa = 10.49KK372 pKa = 8.89EE373 pKa = 4.12LKK375 pKa = 10.59DD376 pKa = 3.45KK377 pKa = 11.0VRR379 pKa = 11.84ANVDD383 pKa = 3.19QMKK386 pKa = 10.62GAFDD390 pKa = 4.66LNLAEE395 pKa = 5.82GGGQLPCEE403 pKa = 4.52SVTIWGRR410 pKa = 11.84SYY412 pKa = 11.07SLCVADD418 pKa = 5.85YY419 pKa = 11.46ADD421 pKa = 3.42QLSNLRR427 pKa = 11.84VALLLMAALIAAFILLRR444 pKa = 11.84DD445 pKa = 3.62
MM1 pKa = 7.79RR2 pKa = 11.84IKK4 pKa = 10.19RR5 pKa = 11.84TLLVLLTLFMSVCASAEE22 pKa = 3.87DD23 pKa = 4.78YY24 pKa = 10.1YY25 pKa = 10.62WPRR28 pKa = 11.84GTQKK32 pKa = 9.82YY33 pKa = 8.53GSYY36 pKa = 9.51MEE38 pKa = 4.29VVEE41 pKa = 4.74EE42 pKa = 3.89ARR44 pKa = 11.84KK45 pKa = 9.19AAIANNPGYY54 pKa = 10.76SRR56 pKa = 11.84VEE58 pKa = 4.01AVRR61 pKa = 11.84VIYY64 pKa = 8.8PANGRR69 pKa = 11.84QDD71 pKa = 3.05MATYY75 pKa = 10.55GLEE78 pKa = 4.63FYY80 pKa = 10.71CLSQGVEE87 pKa = 4.42RR88 pKa = 11.84MCSTSYY94 pKa = 11.27NNPVYY99 pKa = 10.49RR100 pKa = 11.84KK101 pKa = 10.45GEE103 pKa = 3.98GCTAPKK109 pKa = 10.31IPDD112 pKa = 3.7EE113 pKa = 4.32TTGTCKK119 pKa = 10.35EE120 pKa = 4.38PPTPPEE126 pKa = 4.1DD127 pKa = 4.36CIKK130 pKa = 11.06GLTDD134 pKa = 4.65LFSSPPSNIFVSGGRR149 pKa = 11.84NFVNSSPPTGCKK161 pKa = 9.91NGCQYY166 pKa = 11.41LPTTSKK172 pKa = 7.66TTSCYY177 pKa = 10.13RR178 pKa = 11.84YY179 pKa = 9.48PGSDD183 pKa = 2.89NQGFCNYY190 pKa = 9.86VLMTDD195 pKa = 4.32GSACAADD202 pKa = 3.63SGNPGMTGPSLNDD215 pKa = 3.57TPPTNPDD222 pKa = 3.95EE223 pKa = 4.85PPSDD227 pKa = 4.41PNDD230 pKa = 4.17PGCPPGYY237 pKa = 8.96SWSGTTCVKK246 pKa = 10.27TPTDD250 pKa = 3.57PTEE253 pKa = 4.25PGGDD257 pKa = 3.65GGDD260 pKa = 3.91GGDD263 pKa = 4.12GGNTGGGDD271 pKa = 3.29GGGDD275 pKa = 2.96NGGGNDD281 pKa = 3.77NGGGDD286 pKa = 3.94GGTGGSDD293 pKa = 2.98GSGGNGEE300 pKa = 4.28GGGDD304 pKa = 3.57GSGGGDD310 pKa = 3.27GSGGGTGGGDD320 pKa = 3.45GGDD323 pKa = 3.81GGDD326 pKa = 5.45GGNCDD331 pKa = 4.17PAKK334 pKa = 10.16QGCSPGPAGPGGEE347 pKa = 4.21LKK349 pKa = 10.5EE350 pKa = 4.48PKK352 pKa = 9.97PGTWDD357 pKa = 3.36DD358 pKa = 6.11AIATWEE364 pKa = 4.22QKK366 pKa = 10.35VEE368 pKa = 4.01QAKK371 pKa = 10.49KK372 pKa = 8.89EE373 pKa = 4.12LKK375 pKa = 10.59DD376 pKa = 3.45KK377 pKa = 11.0VRR379 pKa = 11.84ANVDD383 pKa = 3.19QMKK386 pKa = 10.62GAFDD390 pKa = 4.66LNLAEE395 pKa = 5.82GGGQLPCEE403 pKa = 4.52SVTIWGRR410 pKa = 11.84SYY412 pKa = 11.07SLCVADD418 pKa = 5.85YY419 pKa = 11.46ADD421 pKa = 3.42QLSNLRR427 pKa = 11.84VALLLMAALIAAFILLRR444 pKa = 11.84DD445 pKa = 3.62
Molecular weight: 45.55 kDa
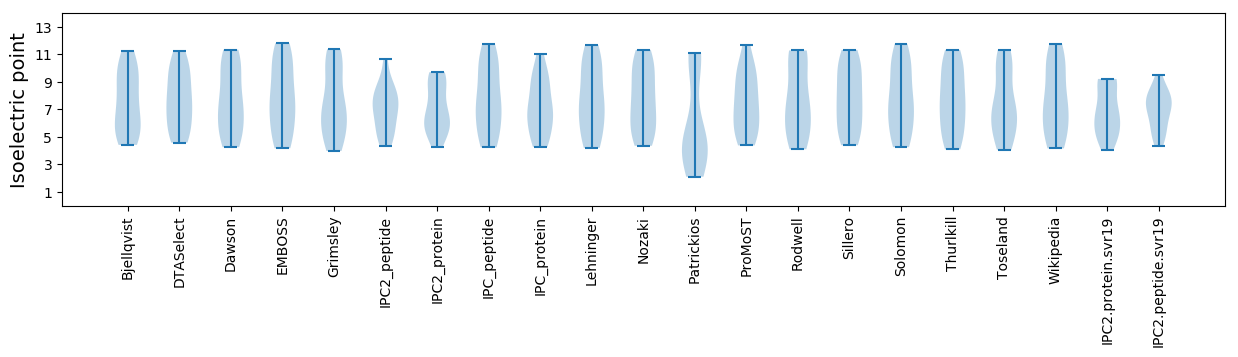
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B9J9N2|A0A6B9J9N2_9VIRU Replication initiation protein OS=Pseudomonas phage pf8_ST274-AUS411 OX=2686285 PE=4 SV=1
MM1 pKa = 7.58LAKK4 pKa = 9.49TLKK7 pKa = 10.76ALLLLCLIQAARR19 pKa = 11.84TVADD23 pKa = 3.95PVKK26 pKa = 10.4GRR28 pKa = 11.84APGSSEE34 pKa = 3.48QLHH37 pKa = 6.03RR38 pKa = 11.84SGEE41 pKa = 4.27RR42 pKa = 11.84KK43 pKa = 9.53HH44 pKa = 6.5GRR46 pKa = 11.84SAPLNASPLKK56 pKa = 10.05QPPLGSVGQLLRR68 pKa = 11.84PALPSPRR75 pKa = 11.84RR76 pKa = 11.84QEE78 pKa = 4.28RR79 pKa = 11.84DD80 pKa = 3.23DD81 pKa = 3.5KK82 pKa = 11.56GRR84 pKa = 11.84ALGVALRR91 pKa = 11.84VPRR94 pKa = 11.84RR95 pKa = 11.84SVPLAVGDD103 pKa = 3.97DD104 pKa = 3.83VAIGTVTRR112 pKa = 11.84MGRR115 pKa = 11.84DD116 pKa = 3.43EE117 pKa = 5.0HH118 pKa = 7.03PWLAWFASEE127 pKa = 4.71
MM1 pKa = 7.58LAKK4 pKa = 9.49TLKK7 pKa = 10.76ALLLLCLIQAARR19 pKa = 11.84TVADD23 pKa = 3.95PVKK26 pKa = 10.4GRR28 pKa = 11.84APGSSEE34 pKa = 3.48QLHH37 pKa = 6.03RR38 pKa = 11.84SGEE41 pKa = 4.27RR42 pKa = 11.84KK43 pKa = 9.53HH44 pKa = 6.5GRR46 pKa = 11.84SAPLNASPLKK56 pKa = 10.05QPPLGSVGQLLRR68 pKa = 11.84PALPSPRR75 pKa = 11.84RR76 pKa = 11.84QEE78 pKa = 4.28RR79 pKa = 11.84DD80 pKa = 3.23DD81 pKa = 3.5KK82 pKa = 11.56GRR84 pKa = 11.84ALGVALRR91 pKa = 11.84VPRR94 pKa = 11.84RR95 pKa = 11.84SVPLAVGDD103 pKa = 3.97DD104 pKa = 3.83VAIGTVTRR112 pKa = 11.84MGRR115 pKa = 11.84DD116 pKa = 3.43EE117 pKa = 5.0HH118 pKa = 7.03PWLAWFASEE127 pKa = 4.71
Molecular weight: 13.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3003 |
30 |
512 |
187.7 |
20.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.724 ± 0.998 | 1.532 ± 0.418 |
5.661 ± 0.422 | 5.128 ± 0.405 |
3.63 ± 0.439 | 8.858 ± 1.695 |
2.198 ± 0.558 | 5.128 ± 0.738 |
4.862 ± 0.438 | 9.491 ± 0.916 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.331 ± 0.24 | 3.064 ± 0.545 |
6.327 ± 0.894 | 3.497 ± 0.297 |
6.593 ± 0.9 | 6.294 ± 0.581 |
5.295 ± 0.509 | 5.794 ± 0.355 |
1.732 ± 0.301 | 2.864 ± 0.317 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
