
Mahlapitsi orthoreovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Orthoreovirus
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
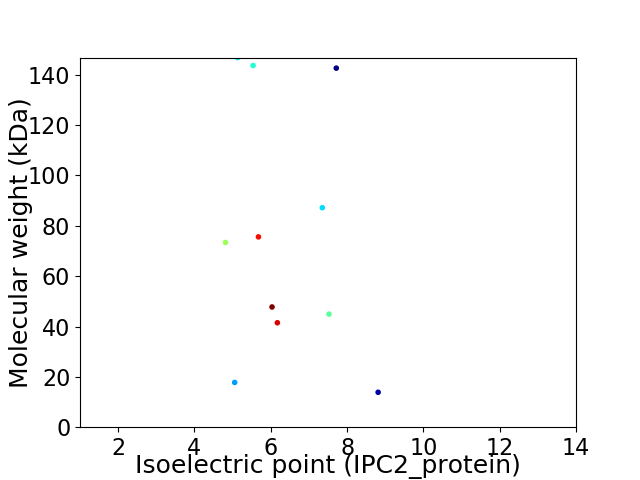
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G1HJT8|A0A3G1HJT8_9REOV Mu B OS=Mahlapitsi orthoreovirus OX=2170064 PE=4 SV=1
MM1 pKa = 6.92GTASSLVQNFNIYY14 pKa = 11.06GNGNVFRR21 pKa = 11.84PTADD25 pKa = 3.15QVSTAVPTLSLQPGVLNPGGLAWRR49 pKa = 11.84RR50 pKa = 11.84INPDD54 pKa = 2.78VDD56 pKa = 3.53PTAPGALIQMTTDD69 pKa = 5.27DD70 pKa = 4.89IPTASLTCTEE80 pKa = 4.28NASGLVPVNAMYY92 pKa = 10.67MMLKK96 pKa = 10.02KK97 pKa = 10.64EE98 pKa = 4.08EE99 pKa = 4.18LVVVTDD105 pKa = 3.63HH106 pKa = 7.7AIDD109 pKa = 3.54QMHH112 pKa = 6.57KK113 pKa = 10.67LEE115 pKa = 4.18YY116 pKa = 11.04AMEE119 pKa = 4.66LSRR122 pKa = 11.84AYY124 pKa = 10.87LNEE127 pKa = 3.8KK128 pKa = 10.38SILPQSTSFDD138 pKa = 3.72SYY140 pKa = 10.97VIRR143 pKa = 11.84VDD145 pKa = 3.9CYY147 pKa = 9.95VGASALQAARR157 pKa = 11.84NFVRR161 pKa = 11.84GAPLITRR168 pKa = 11.84SRR170 pKa = 11.84MVAYY174 pKa = 7.45MTAIQNAFEE183 pKa = 4.83ILSDD187 pKa = 3.51WEE189 pKa = 4.07SDD191 pKa = 2.7IRR193 pKa = 11.84EE194 pKa = 4.25AMNLIPSTTPYY205 pKa = 11.08GATTCDD211 pKa = 3.33MQSVIKK217 pKa = 10.39FLDD220 pKa = 3.44EE221 pKa = 4.11NLPEE225 pKa = 4.29NSLVRR230 pKa = 11.84LYY232 pKa = 9.55PQQAADD238 pKa = 3.64SIARR242 pKa = 11.84RR243 pKa = 11.84NGGIRR248 pKa = 11.84WKK250 pKa = 11.01DD251 pKa = 3.57EE252 pKa = 3.77VTGDD256 pKa = 3.81VPSLATNTVAAATLGVINNTVPLGEE281 pKa = 4.56KK282 pKa = 10.32SDD284 pKa = 3.84VTSEE288 pKa = 3.81AMSLVTVVKK297 pKa = 9.88PDD299 pKa = 3.55LVTSMRR305 pKa = 11.84PVSSSVFATTVSPKK319 pKa = 9.46TYY321 pKa = 10.2NIRR324 pKa = 11.84AQPVMEE330 pKa = 4.53ALWLRR335 pKa = 11.84TAAAGVDD342 pKa = 3.79MVVGWWSEE350 pKa = 3.78EE351 pKa = 3.81HH352 pKa = 6.73AATHH356 pKa = 5.05YY357 pKa = 9.83TVIRR361 pKa = 11.84PGTKK365 pKa = 9.92VINLQQTGSMLITVDD380 pKa = 5.74LVDD383 pKa = 3.29KK384 pKa = 10.58DD385 pKa = 3.48YY386 pKa = 10.62RR387 pKa = 11.84TVHH390 pKa = 6.36FNPDD394 pKa = 3.25GQTVILLLLQSKK406 pKa = 10.33IPFEE410 pKa = 4.11QLTSPTEE417 pKa = 4.38LIGVAQVASVVLSASDD433 pKa = 3.42SSTEE437 pKa = 3.67GSAIINNTNLSYY449 pKa = 10.97LFEE452 pKa = 5.4RR453 pKa = 11.84EE454 pKa = 3.89ILRR457 pKa = 11.84GPDD460 pKa = 3.45TEE462 pKa = 4.42VNTYY466 pKa = 9.45LLCGFRR472 pKa = 11.84TDD474 pKa = 3.78SQPTKK479 pKa = 11.09EE480 pKa = 4.01NMPWYY485 pKa = 9.58DD486 pKa = 2.9AWDD489 pKa = 3.72AKK491 pKa = 8.49TTLTPLTTGEE501 pKa = 4.17VTLNGAAVPFVTPMSLIGAYY521 pKa = 8.56TPEE524 pKa = 4.15AMQGALPNDD533 pKa = 3.63AGILLAEE540 pKa = 4.3RR541 pKa = 11.84AMNLAEE547 pKa = 4.85AIKK550 pKa = 11.07LEE552 pKa = 4.61DD553 pKa = 4.7DD554 pKa = 3.87SMADD558 pKa = 3.32EE559 pKa = 5.2ASPFSAPIQGVLAIQQHH576 pKa = 5.32EE577 pKa = 4.18PGEE580 pKa = 4.47GVKK583 pKa = 10.39AWLGPFDD590 pKa = 4.53ILRR593 pKa = 11.84KK594 pKa = 8.85AARR597 pKa = 11.84AIQVFLGNPKK607 pKa = 9.79SVLMQGVPVVSDD619 pKa = 3.89PNVWIAMVQGIRR631 pKa = 11.84DD632 pKa = 4.59GIRR635 pKa = 11.84NKK637 pKa = 10.41SLSVGVRR644 pKa = 11.84TALDD648 pKa = 3.4NLNAVQSVRR657 pKa = 11.84TWKK660 pKa = 10.56SDD662 pKa = 3.16FLTKK666 pKa = 10.22VEE668 pKa = 4.49KK669 pKa = 10.64YY670 pKa = 9.97YY671 pKa = 10.45PPSSEE676 pKa = 3.81
MM1 pKa = 6.92GTASSLVQNFNIYY14 pKa = 11.06GNGNVFRR21 pKa = 11.84PTADD25 pKa = 3.15QVSTAVPTLSLQPGVLNPGGLAWRR49 pKa = 11.84RR50 pKa = 11.84INPDD54 pKa = 2.78VDD56 pKa = 3.53PTAPGALIQMTTDD69 pKa = 5.27DD70 pKa = 4.89IPTASLTCTEE80 pKa = 4.28NASGLVPVNAMYY92 pKa = 10.67MMLKK96 pKa = 10.02KK97 pKa = 10.64EE98 pKa = 4.08EE99 pKa = 4.18LVVVTDD105 pKa = 3.63HH106 pKa = 7.7AIDD109 pKa = 3.54QMHH112 pKa = 6.57KK113 pKa = 10.67LEE115 pKa = 4.18YY116 pKa = 11.04AMEE119 pKa = 4.66LSRR122 pKa = 11.84AYY124 pKa = 10.87LNEE127 pKa = 3.8KK128 pKa = 10.38SILPQSTSFDD138 pKa = 3.72SYY140 pKa = 10.97VIRR143 pKa = 11.84VDD145 pKa = 3.9CYY147 pKa = 9.95VGASALQAARR157 pKa = 11.84NFVRR161 pKa = 11.84GAPLITRR168 pKa = 11.84SRR170 pKa = 11.84MVAYY174 pKa = 7.45MTAIQNAFEE183 pKa = 4.83ILSDD187 pKa = 3.51WEE189 pKa = 4.07SDD191 pKa = 2.7IRR193 pKa = 11.84EE194 pKa = 4.25AMNLIPSTTPYY205 pKa = 11.08GATTCDD211 pKa = 3.33MQSVIKK217 pKa = 10.39FLDD220 pKa = 3.44EE221 pKa = 4.11NLPEE225 pKa = 4.29NSLVRR230 pKa = 11.84LYY232 pKa = 9.55PQQAADD238 pKa = 3.64SIARR242 pKa = 11.84RR243 pKa = 11.84NGGIRR248 pKa = 11.84WKK250 pKa = 11.01DD251 pKa = 3.57EE252 pKa = 3.77VTGDD256 pKa = 3.81VPSLATNTVAAATLGVINNTVPLGEE281 pKa = 4.56KK282 pKa = 10.32SDD284 pKa = 3.84VTSEE288 pKa = 3.81AMSLVTVVKK297 pKa = 9.88PDD299 pKa = 3.55LVTSMRR305 pKa = 11.84PVSSSVFATTVSPKK319 pKa = 9.46TYY321 pKa = 10.2NIRR324 pKa = 11.84AQPVMEE330 pKa = 4.53ALWLRR335 pKa = 11.84TAAAGVDD342 pKa = 3.79MVVGWWSEE350 pKa = 3.78EE351 pKa = 3.81HH352 pKa = 6.73AATHH356 pKa = 5.05YY357 pKa = 9.83TVIRR361 pKa = 11.84PGTKK365 pKa = 9.92VINLQQTGSMLITVDD380 pKa = 5.74LVDD383 pKa = 3.29KK384 pKa = 10.58DD385 pKa = 3.48YY386 pKa = 10.62RR387 pKa = 11.84TVHH390 pKa = 6.36FNPDD394 pKa = 3.25GQTVILLLLQSKK406 pKa = 10.33IPFEE410 pKa = 4.11QLTSPTEE417 pKa = 4.38LIGVAQVASVVLSASDD433 pKa = 3.42SSTEE437 pKa = 3.67GSAIINNTNLSYY449 pKa = 10.97LFEE452 pKa = 5.4RR453 pKa = 11.84EE454 pKa = 3.89ILRR457 pKa = 11.84GPDD460 pKa = 3.45TEE462 pKa = 4.42VNTYY466 pKa = 9.45LLCGFRR472 pKa = 11.84TDD474 pKa = 3.78SQPTKK479 pKa = 11.09EE480 pKa = 4.01NMPWYY485 pKa = 9.58DD486 pKa = 2.9AWDD489 pKa = 3.72AKK491 pKa = 8.49TTLTPLTTGEE501 pKa = 4.17VTLNGAAVPFVTPMSLIGAYY521 pKa = 8.56TPEE524 pKa = 4.15AMQGALPNDD533 pKa = 3.63AGILLAEE540 pKa = 4.3RR541 pKa = 11.84AMNLAEE547 pKa = 4.85AIKK550 pKa = 11.07LEE552 pKa = 4.61DD553 pKa = 4.7DD554 pKa = 3.87SMADD558 pKa = 3.32EE559 pKa = 5.2ASPFSAPIQGVLAIQQHH576 pKa = 5.32EE577 pKa = 4.18PGEE580 pKa = 4.47GVKK583 pKa = 10.39AWLGPFDD590 pKa = 4.53ILRR593 pKa = 11.84KK594 pKa = 8.85AARR597 pKa = 11.84AIQVFLGNPKK607 pKa = 9.79SVLMQGVPVVSDD619 pKa = 3.89PNVWIAMVQGIRR631 pKa = 11.84DD632 pKa = 4.59GIRR635 pKa = 11.84NKK637 pKa = 10.41SLSVGVRR644 pKa = 11.84TALDD648 pKa = 3.4NLNAVQSVRR657 pKa = 11.84TWKK660 pKa = 10.56SDD662 pKa = 3.16FLTKK666 pKa = 10.22VEE668 pKa = 4.49KK669 pKa = 10.64YY670 pKa = 9.97YY671 pKa = 10.45PPSSEE676 pKa = 3.81
Molecular weight: 73.4 kDa
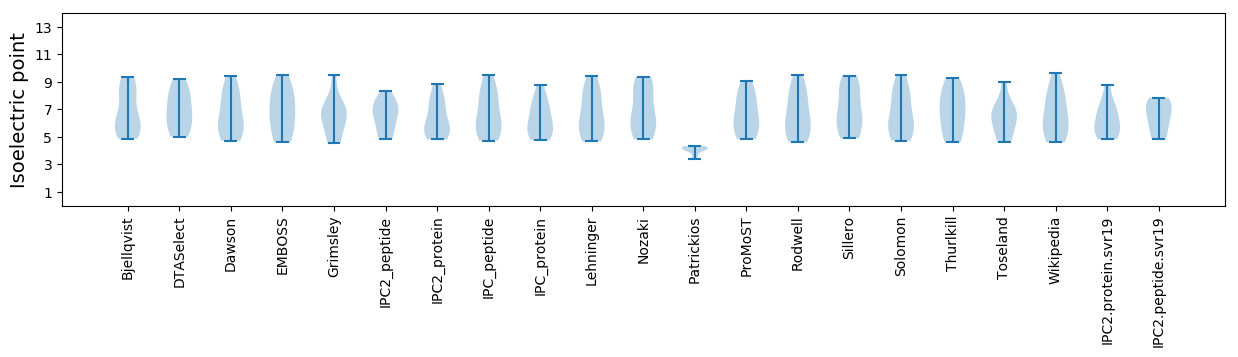
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G1HJT8|A0A3G1HJT8_9REOV Mu B OS=Mahlapitsi orthoreovirus OX=2170064 PE=4 SV=1
MM1 pKa = 7.64GSGPSNFVNHH11 pKa = 6.97ASGEE15 pKa = 4.21AIISGLSDD23 pKa = 2.93QTNRR27 pKa = 11.84LGSLLSQNVYY37 pKa = 10.35NIIYY41 pKa = 8.74FFVIGGLILSAGYY54 pKa = 10.54GLYY57 pKa = 10.1KK58 pKa = 9.18YY59 pKa = 10.27CKK61 pKa = 8.23YY62 pKa = 10.01KK63 pKa = 10.28QRR65 pKa = 11.84RR66 pKa = 11.84AKK68 pKa = 10.58NLTRR72 pKa = 11.84QLSRR76 pKa = 11.84EE77 pKa = 4.15LIDD80 pKa = 3.85LNRR83 pKa = 11.84KK84 pKa = 8.84IEE86 pKa = 4.62HH87 pKa = 6.47ISGGKK92 pKa = 9.28IPATKK97 pKa = 9.9PSAPRR102 pKa = 11.84YY103 pKa = 7.11TPPCYY108 pKa = 9.97KK109 pKa = 10.33EE110 pKa = 4.03PIYY113 pKa = 11.27NEE115 pKa = 3.82VCEE118 pKa = 4.35GGFYY122 pKa = 10.94GNCC125 pKa = 3.45
MM1 pKa = 7.64GSGPSNFVNHH11 pKa = 6.97ASGEE15 pKa = 4.21AIISGLSDD23 pKa = 2.93QTNRR27 pKa = 11.84LGSLLSQNVYY37 pKa = 10.35NIIYY41 pKa = 8.74FFVIGGLILSAGYY54 pKa = 10.54GLYY57 pKa = 10.1KK58 pKa = 9.18YY59 pKa = 10.27CKK61 pKa = 8.23YY62 pKa = 10.01KK63 pKa = 10.28QRR65 pKa = 11.84RR66 pKa = 11.84AKK68 pKa = 10.58NLTRR72 pKa = 11.84QLSRR76 pKa = 11.84EE77 pKa = 4.15LIDD80 pKa = 3.85LNRR83 pKa = 11.84KK84 pKa = 8.84IEE86 pKa = 4.62HH87 pKa = 6.47ISGGKK92 pKa = 9.28IPATKK97 pKa = 9.9PSAPRR102 pKa = 11.84YY103 pKa = 7.11TPPCYY108 pKa = 9.97KK109 pKa = 10.33EE110 pKa = 4.03PIYY113 pKa = 11.27NEE115 pKa = 3.82VCEE118 pKa = 4.35GGFYY122 pKa = 10.94GNCC125 pKa = 3.45
Molecular weight: 13.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7411 |
125 |
1294 |
673.7 |
75.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.597 ± 0.374 | 1.336 ± 0.126 |
5.937 ± 0.211 | 4.723 ± 0.302 |
3.562 ± 0.304 | 4.844 ± 0.258 |
2.375 ± 0.212 | 5.667 ± 0.257 |
3.967 ± 0.377 | 9.702 ± 0.317 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.996 ± 0.162 | 5.101 ± 0.33 |
5.735 ± 0.195 | 4.089 ± 0.213 |
5.735 ± 0.244 | 6.99 ± 0.236 |
6.207 ± 0.27 | 7.786 ± 0.359 |
1.741 ± 0.162 | 3.913 ± 0.357 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
