
Pseudohoeflea suaedae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Pseudohoeflea
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
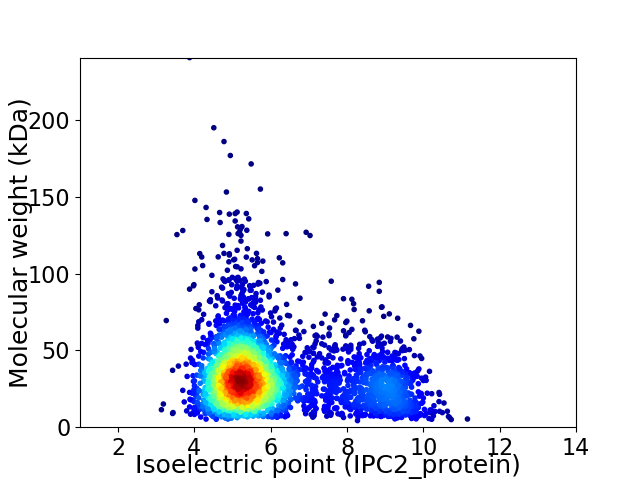
Virtual 2D-PAGE plot for 3469 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V3A6X2|A0A4V3A6X2_9RHIZ Carbohydrate kinase OS=Pseudohoeflea suaedae OX=877384 GN=E2A64_14740 PE=3 SV=1
MM1 pKa = 7.67RR2 pKa = 11.84SVILFASFVLALVSSCFLLSSNSHH26 pKa = 6.57AAPFPCEE33 pKa = 3.8GNTISSGNEE42 pKa = 3.07IVVQLDD48 pKa = 3.53GAITCGTAYY57 pKa = 10.86NSTDD61 pKa = 3.51EE62 pKa = 5.6IEE64 pKa = 4.14ILNNGIVSGSTIQVLIAFYY83 pKa = 10.83NGATGSVTGGTTSDD97 pKa = 3.67DD98 pKa = 3.44SGTFLSPEE106 pKa = 4.16CNVTVGCTLSVTGTYY121 pKa = 10.4SGTPFAFSVVFPGANSTTATLSNTNYY147 pKa = 10.22GAATAPEE154 pKa = 4.11ISVSEE159 pKa = 4.1TGQNQGAVADD169 pKa = 4.67EE170 pKa = 4.38GTLAQGTQAAGSPVNLTFTVTNSGTGDD197 pKa = 3.54LTIATATSSLLSNVTVNSISAPTNTTVASGGGTEE231 pKa = 4.43TFTVQYY237 pKa = 9.74TPTSAGNFSFNLSFANDD254 pKa = 3.95DD255 pKa = 4.14ANEE258 pKa = 4.2GPYY261 pKa = 10.93NFIVSGTASGAPEE274 pKa = 3.68ISVAEE279 pKa = 4.27TGQSQGAVADD289 pKa = 4.42EE290 pKa = 4.38GTLAQGTQAAGSPVNLTFTVTNSGTGDD317 pKa = 3.54LTIATATSSLLSNVTVNSISAPTNSTVASGGGTEE351 pKa = 4.43TFTVQYY357 pKa = 9.74TPTSAGNFSFNLSFVNDD374 pKa = 4.35DD375 pKa = 4.29ANEE378 pKa = 4.0SPYY381 pKa = 11.27NFTISGTGTAAPTFSQSFSPTTIEE405 pKa = 3.62IGGASTITFTIDD417 pKa = 2.69NSTNALDD424 pKa = 3.79ATSLDD429 pKa = 3.87FTDD432 pKa = 4.42NLPAGMNVSSPSNAATTCTGGTLTASSGAGTVSYY466 pKa = 11.05SGGTVGAGASCTVSVDD482 pKa = 3.29VGATSDD488 pKa = 3.42GSFVNTTGNLTSSLGNSGPSSASLTVSSPDD518 pKa = 2.91IDD520 pKa = 3.72VQRR523 pKa = 11.84PVSTSIADD531 pKa = 4.26DD532 pKa = 5.4DD533 pKa = 4.33IDD535 pKa = 4.01SQGVVTLGVQQTLTFTIEE553 pKa = 4.2NKK555 pKa = 8.8GTATLSLSGTPTSDD569 pKa = 2.9SPINVSVDD577 pKa = 3.79SISAPGSNTLAGGATTTFTVQYY599 pKa = 9.02TPIAIGPFSFQVDD612 pKa = 3.56IVSNDD617 pKa = 3.17ADD619 pKa = 3.67EE620 pKa = 4.76ANYY623 pKa = 10.51DD624 pKa = 3.62ILVSGTASGAAEE636 pKa = 4.04IEE638 pKa = 4.52VSSSASGPISDD649 pKa = 5.1GGTDD653 pKa = 3.87TISGTVTAGSSATITYY669 pKa = 7.78TITNSGTAPLSLTQPTVGGNVTSTTNVTVDD699 pKa = 3.22SFTLSSLSVVSGGGTATLQVSYY721 pKa = 9.79TPTSSGSFDD730 pKa = 3.89FDD732 pKa = 4.99FNFANNDD739 pKa = 3.14ADD741 pKa = 3.94EE742 pKa = 4.6NPFNISVSGTGGGIPAGLSATSGSGQSTEE771 pKa = 4.48AGTQFGSVLVATVTDD786 pKa = 3.73SSNAGVAGVSVTFTAPSSGASLTFAGSGSNTEE818 pKa = 4.26TVITASDD825 pKa = 3.66GTATSSAMTANAIASGYY842 pKa = 10.38SGGSLQSYY850 pKa = 10.04AVTASAAGLTSVDD863 pKa = 4.33FFLTNSRR870 pKa = 11.84DD871 pKa = 3.45SAADD875 pKa = 3.19IQRR878 pKa = 11.84TEE880 pKa = 3.99DD881 pKa = 3.51VIASFVTSRR890 pKa = 11.84ANVIVAGQPDD900 pKa = 4.67LVSRR904 pKa = 11.84LTNGPFGRR912 pKa = 11.84QSGLNGFNFNASRR925 pKa = 11.84FSQTGSFQFSLRR937 pKa = 11.84AFSDD941 pKa = 3.42AVRR944 pKa = 11.84NGVSGSPVHH953 pKa = 6.2QPVTAAGHH961 pKa = 6.0AAGFDD966 pKa = 3.73SFTSEE971 pKa = 4.09TTSGYY976 pKa = 11.3ASAAPQAAEE985 pKa = 4.25AEE987 pKa = 4.41TLAILGIAAGQAPAAGEE1004 pKa = 3.96EE1005 pKa = 4.36SAEE1008 pKa = 4.27VQSGWDD1014 pKa = 3.67FWAEE1018 pKa = 3.5GTYY1021 pKa = 11.15ARR1023 pKa = 11.84TQDD1026 pKa = 3.46KK1027 pKa = 11.15ASDD1030 pKa = 3.93SVSGLFFAGLDD1041 pKa = 3.34YY1042 pKa = 11.09RR1043 pKa = 11.84FADD1046 pKa = 3.88RR1047 pKa = 11.84AVVGVLGQLDD1057 pKa = 3.5ITDD1060 pKa = 3.78EE1061 pKa = 4.62HH1062 pKa = 6.79NGSANTSADD1071 pKa = 3.68GIGWMVGPYY1080 pKa = 10.37AVLKK1084 pKa = 8.75VHH1086 pKa = 6.06QNFYY1090 pKa = 10.66IDD1092 pKa = 3.56GAVTYY1097 pKa = 8.58GQSNNSVNALGLFEE1111 pKa = 6.75DD1112 pKa = 4.66DD1113 pKa = 4.71FRR1115 pKa = 11.84TQRR1118 pKa = 11.84FLMQGGLTGDD1128 pKa = 3.99FEE1130 pKa = 5.54IDD1132 pKa = 2.72EE1133 pKa = 4.66HH1134 pKa = 8.56ARR1136 pKa = 11.84ISPFVRR1142 pKa = 11.84LTYY1145 pKa = 10.32YY1146 pKa = 10.74YY1147 pKa = 10.09EE1148 pKa = 4.16EE1149 pKa = 4.32QEE1151 pKa = 4.64SYY1153 pKa = 10.89TDD1155 pKa = 3.4SLGNQIPSQDD1165 pKa = 3.19FDD1167 pKa = 3.83LGRR1170 pKa = 11.84VEE1172 pKa = 5.2FGPKK1176 pKa = 9.76ISWEE1180 pKa = 4.36LILDD1184 pKa = 4.07DD1185 pKa = 5.25SLLFSPFVSLSGIYY1199 pKa = 10.23DD1200 pKa = 3.5FNKK1203 pKa = 10.04LHH1205 pKa = 6.08EE1206 pKa = 4.64TVPTDD1211 pKa = 3.34PRR1213 pKa = 11.84LASSDD1218 pKa = 3.19EE1219 pKa = 3.97DD1220 pKa = 3.31LRR1222 pKa = 11.84ARR1224 pKa = 11.84LEE1226 pKa = 3.9MGAGIVIPNRR1236 pKa = 11.84KK1237 pKa = 8.73IRR1239 pKa = 11.84ISGKK1243 pKa = 10.14GFYY1246 pKa = 10.29DD1247 pKa = 4.71GIGAADD1253 pKa = 4.68FSSYY1257 pKa = 11.26GGTLNFTVPFF1267 pKa = 4.53
MM1 pKa = 7.67RR2 pKa = 11.84SVILFASFVLALVSSCFLLSSNSHH26 pKa = 6.57AAPFPCEE33 pKa = 3.8GNTISSGNEE42 pKa = 3.07IVVQLDD48 pKa = 3.53GAITCGTAYY57 pKa = 10.86NSTDD61 pKa = 3.51EE62 pKa = 5.6IEE64 pKa = 4.14ILNNGIVSGSTIQVLIAFYY83 pKa = 10.83NGATGSVTGGTTSDD97 pKa = 3.67DD98 pKa = 3.44SGTFLSPEE106 pKa = 4.16CNVTVGCTLSVTGTYY121 pKa = 10.4SGTPFAFSVVFPGANSTTATLSNTNYY147 pKa = 10.22GAATAPEE154 pKa = 4.11ISVSEE159 pKa = 4.1TGQNQGAVADD169 pKa = 4.67EE170 pKa = 4.38GTLAQGTQAAGSPVNLTFTVTNSGTGDD197 pKa = 3.54LTIATATSSLLSNVTVNSISAPTNTTVASGGGTEE231 pKa = 4.43TFTVQYY237 pKa = 9.74TPTSAGNFSFNLSFANDD254 pKa = 3.95DD255 pKa = 4.14ANEE258 pKa = 4.2GPYY261 pKa = 10.93NFIVSGTASGAPEE274 pKa = 3.68ISVAEE279 pKa = 4.27TGQSQGAVADD289 pKa = 4.42EE290 pKa = 4.38GTLAQGTQAAGSPVNLTFTVTNSGTGDD317 pKa = 3.54LTIATATSSLLSNVTVNSISAPTNSTVASGGGTEE351 pKa = 4.43TFTVQYY357 pKa = 9.74TPTSAGNFSFNLSFVNDD374 pKa = 4.35DD375 pKa = 4.29ANEE378 pKa = 4.0SPYY381 pKa = 11.27NFTISGTGTAAPTFSQSFSPTTIEE405 pKa = 3.62IGGASTITFTIDD417 pKa = 2.69NSTNALDD424 pKa = 3.79ATSLDD429 pKa = 3.87FTDD432 pKa = 4.42NLPAGMNVSSPSNAATTCTGGTLTASSGAGTVSYY466 pKa = 11.05SGGTVGAGASCTVSVDD482 pKa = 3.29VGATSDD488 pKa = 3.42GSFVNTTGNLTSSLGNSGPSSASLTVSSPDD518 pKa = 2.91IDD520 pKa = 3.72VQRR523 pKa = 11.84PVSTSIADD531 pKa = 4.26DD532 pKa = 5.4DD533 pKa = 4.33IDD535 pKa = 4.01SQGVVTLGVQQTLTFTIEE553 pKa = 4.2NKK555 pKa = 8.8GTATLSLSGTPTSDD569 pKa = 2.9SPINVSVDD577 pKa = 3.79SISAPGSNTLAGGATTTFTVQYY599 pKa = 9.02TPIAIGPFSFQVDD612 pKa = 3.56IVSNDD617 pKa = 3.17ADD619 pKa = 3.67EE620 pKa = 4.76ANYY623 pKa = 10.51DD624 pKa = 3.62ILVSGTASGAAEE636 pKa = 4.04IEE638 pKa = 4.52VSSSASGPISDD649 pKa = 5.1GGTDD653 pKa = 3.87TISGTVTAGSSATITYY669 pKa = 7.78TITNSGTAPLSLTQPTVGGNVTSTTNVTVDD699 pKa = 3.22SFTLSSLSVVSGGGTATLQVSYY721 pKa = 9.79TPTSSGSFDD730 pKa = 3.89FDD732 pKa = 4.99FNFANNDD739 pKa = 3.14ADD741 pKa = 3.94EE742 pKa = 4.6NPFNISVSGTGGGIPAGLSATSGSGQSTEE771 pKa = 4.48AGTQFGSVLVATVTDD786 pKa = 3.73SSNAGVAGVSVTFTAPSSGASLTFAGSGSNTEE818 pKa = 4.26TVITASDD825 pKa = 3.66GTATSSAMTANAIASGYY842 pKa = 10.38SGGSLQSYY850 pKa = 10.04AVTASAAGLTSVDD863 pKa = 4.33FFLTNSRR870 pKa = 11.84DD871 pKa = 3.45SAADD875 pKa = 3.19IQRR878 pKa = 11.84TEE880 pKa = 3.99DD881 pKa = 3.51VIASFVTSRR890 pKa = 11.84ANVIVAGQPDD900 pKa = 4.67LVSRR904 pKa = 11.84LTNGPFGRR912 pKa = 11.84QSGLNGFNFNASRR925 pKa = 11.84FSQTGSFQFSLRR937 pKa = 11.84AFSDD941 pKa = 3.42AVRR944 pKa = 11.84NGVSGSPVHH953 pKa = 6.2QPVTAAGHH961 pKa = 6.0AAGFDD966 pKa = 3.73SFTSEE971 pKa = 4.09TTSGYY976 pKa = 11.3ASAAPQAAEE985 pKa = 4.25AEE987 pKa = 4.41TLAILGIAAGQAPAAGEE1004 pKa = 3.96EE1005 pKa = 4.36SAEE1008 pKa = 4.27VQSGWDD1014 pKa = 3.67FWAEE1018 pKa = 3.5GTYY1021 pKa = 11.15ARR1023 pKa = 11.84TQDD1026 pKa = 3.46KK1027 pKa = 11.15ASDD1030 pKa = 3.93SVSGLFFAGLDD1041 pKa = 3.34YY1042 pKa = 11.09RR1043 pKa = 11.84FADD1046 pKa = 3.88RR1047 pKa = 11.84AVVGVLGQLDD1057 pKa = 3.5ITDD1060 pKa = 3.78EE1061 pKa = 4.62HH1062 pKa = 6.79NGSANTSADD1071 pKa = 3.68GIGWMVGPYY1080 pKa = 10.37AVLKK1084 pKa = 8.75VHH1086 pKa = 6.06QNFYY1090 pKa = 10.66IDD1092 pKa = 3.56GAVTYY1097 pKa = 8.58GQSNNSVNALGLFEE1111 pKa = 6.75DD1112 pKa = 4.66DD1113 pKa = 4.71FRR1115 pKa = 11.84TQRR1118 pKa = 11.84FLMQGGLTGDD1128 pKa = 3.99FEE1130 pKa = 5.54IDD1132 pKa = 2.72EE1133 pKa = 4.66HH1134 pKa = 8.56ARR1136 pKa = 11.84ISPFVRR1142 pKa = 11.84LTYY1145 pKa = 10.32YY1146 pKa = 10.74YY1147 pKa = 10.09EE1148 pKa = 4.16EE1149 pKa = 4.32QEE1151 pKa = 4.64SYY1153 pKa = 10.89TDD1155 pKa = 3.4SLGNQIPSQDD1165 pKa = 3.19FDD1167 pKa = 3.83LGRR1170 pKa = 11.84VEE1172 pKa = 5.2FGPKK1176 pKa = 9.76ISWEE1180 pKa = 4.36LILDD1184 pKa = 4.07DD1185 pKa = 5.25SLLFSPFVSLSGIYY1199 pKa = 10.23DD1200 pKa = 3.5FNKK1203 pKa = 10.04LHH1205 pKa = 6.08EE1206 pKa = 4.64TVPTDD1211 pKa = 3.34PRR1213 pKa = 11.84LASSDD1218 pKa = 3.19EE1219 pKa = 3.97DD1220 pKa = 3.31LRR1222 pKa = 11.84ARR1224 pKa = 11.84LEE1226 pKa = 3.9MGAGIVIPNRR1236 pKa = 11.84KK1237 pKa = 8.73IRR1239 pKa = 11.84ISGKK1243 pKa = 10.14GFYY1246 pKa = 10.29DD1247 pKa = 4.71GIGAADD1253 pKa = 4.68FSSYY1257 pKa = 11.26GGTLNFTVPFF1267 pKa = 4.53
Molecular weight: 128.11 kDa
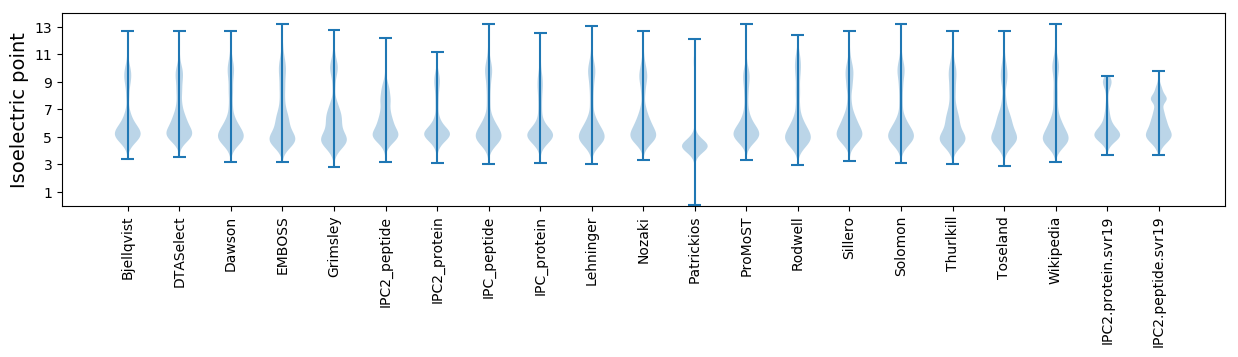
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R5PM99|A0A4R5PM99_9RHIZ XRE family transcriptional regulator OS=Pseudohoeflea suaedae OX=877384 GN=E2A64_02995 PE=4 SV=1
MM1 pKa = 7.62TKK3 pKa = 9.11RR4 pKa = 11.84TYY6 pKa = 10.36QPSKK10 pKa = 9.73LVRR13 pKa = 11.84KK14 pKa = 8.95RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.63GFRR20 pKa = 11.84SRR22 pKa = 11.84MAKK25 pKa = 9.42VGGRR29 pKa = 11.84KK30 pKa = 9.34VIAARR35 pKa = 11.84RR36 pKa = 11.84NRR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.03RR42 pKa = 11.84LSAA45 pKa = 4.03
MM1 pKa = 7.62TKK3 pKa = 9.11RR4 pKa = 11.84TYY6 pKa = 10.36QPSKK10 pKa = 9.73LVRR13 pKa = 11.84KK14 pKa = 8.95RR15 pKa = 11.84RR16 pKa = 11.84HH17 pKa = 4.63GFRR20 pKa = 11.84SRR22 pKa = 11.84MAKK25 pKa = 9.42VGGRR29 pKa = 11.84KK30 pKa = 9.34VIAARR35 pKa = 11.84RR36 pKa = 11.84NRR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.03RR42 pKa = 11.84LSAA45 pKa = 4.03
Molecular weight: 5.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1121147 |
41 |
2560 |
323.2 |
35.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.854 ± 0.06 | 0.81 ± 0.013 |
6.054 ± 0.038 | 6.281 ± 0.041 |
3.892 ± 0.027 | 8.751 ± 0.05 |
1.982 ± 0.02 | 5.67 ± 0.032 |
3.549 ± 0.032 | 9.516 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.691 ± 0.022 | 2.792 ± 0.022 |
4.811 ± 0.031 | 2.802 ± 0.022 |
6.71 ± 0.046 | 5.732 ± 0.03 |
5.346 ± 0.027 | 7.249 ± 0.033 |
1.233 ± 0.015 | 2.274 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
