
Esox lucius (Northern pike)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Protacanthopterygii; Esociformes; Esocidae; Esox
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
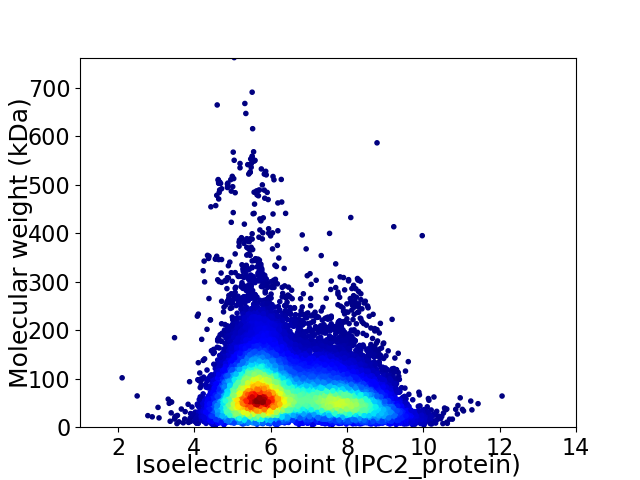
Virtual 2D-PAGE plot for 71519 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6Q2X6C8|A0A6Q2X6C8_ESOLU Multiple C2 and transmembrane domain-containing protein 1 OS=Esox lucius OX=8010 PE=4 SV=1
HHH2 pKa = 7.77LYYY5 pKa = 11.04RR6 pKa = 11.84DDD8 pKa = 3.41KK9 pKa = 10.26QIKKK13 pKa = 7.51KKK15 pKa = 9.87QLGSVRR21 pKa = 11.84CHHH24 pKa = 7.43EEE26 pKa = 3.54DD27 pKa = 4.05DD28 pKa = 3.51CLVDDD33 pKa = 5.31DD34 pKa = 4.66VCFRR38 pKa = 11.84EEE40 pKa = 4.78DD41 pKa = 3.55QCLVDDD47 pKa = 4.72DD48 pKa = 4.45VCFRR52 pKa = 11.84EEE54 pKa = 4.46DD55 pKa = 3.26QSLVDDD61 pKa = 4.34DD62 pKa = 4.79VCFRR66 pKa = 11.84EEE68 pKa = 4.88DD69 pKa = 3.72QCLVDDD75 pKa = 4.72DD76 pKa = 4.45VCFRR80 pKa = 11.84EEE82 pKa = 4.46DD83 pKa = 3.17QSLVDDD89 pKa = 3.49EE90 pKa = 4.91VCFRR94 pKa = 11.84EEE96 pKa = 4.84DD97 pKa = 3.51QSLLDDD103 pKa = 3.55EE104 pKa = 4.59VCFRR108 pKa = 11.84EEE110 pKa = 4.67DD111 pKa = 3.36QSLVDDD117 pKa = 3.49EE118 pKa = 4.91VCFRR122 pKa = 11.84EEE124 pKa = 4.67DD125 pKa = 3.36QSLVDDD131 pKa = 3.49EE132 pKa = 4.91VCFRR136 pKa = 11.84EEE138 pKa = 4.67DD139 pKa = 3.36QSLVDDD145 pKa = 3.49EE146 pKa = 4.91VCFRR150 pKa = 11.84EEE152 pKa = 5.01DD153 pKa = 3.64QCLVDDD159 pKa = 3.71EE160 pKa = 5.03VCFRR164 pKa = 11.84EEE166 pKa = 5.01DD167 pKa = 3.73QCLVDDD173 pKa = 4.95DD174 pKa = 4.92VCFRR178 pKa = 11.84EEE180 pKa = 4.57DD181 pKa = 3.44QSLVDDD187 pKa = 5.05DD188 pKa = 4.6EE189 pKa = 4.78CLRR192 pKa = 11.84EEE194 pKa = 4.48GQCLVDDD201 pKa = 3.38EE202 pKa = 5.04VCFRR206 pKa = 11.84EEE208 pKa = 5.07GQSLVDDD215 pKa = 3.32EE216 pKa = 4.92VCFRR220 pKa = 11.84EEE222 pKa = 4.67DD223 pKa = 3.45QSLVDDD229 pKa = 4.34DD230 pKa = 4.79VCFRR234 pKa = 11.84EEE236 pKa = 4.57DD237 pKa = 3.44QSLVDDD243 pKa = 3.95DD244 pKa = 4.24VCFRR248 pKa = 11.84EEE250 pKa = 4.62GQCLVDDD257 pKa = 3.38EE258 pKa = 5.05VCFRR262 pKa = 11.84EEE264 pKa = 4.67DD265 pKa = 3.45QSLVDDD271 pKa = 4.34DD272 pKa = 4.79VCFRR276 pKa = 11.84EEE278 pKa = 4.88DD279 pKa = 3.72QCLVDDD285 pKa = 4.95DD286 pKa = 4.92VCFRR290 pKa = 11.84EEE292 pKa = 4.88DD293 pKa = 3.72QCLVDDD299 pKa = 4.95DD300 pKa = 4.92VCFRR304 pKa = 11.84EEE306 pKa = 4.88DD307 pKa = 3.72QCLVDDD313 pKa = 4.95DD314 pKa = 4.92VCFRR318 pKa = 11.84EEE320 pKa = 4.88DD321 pKa = 3.72QCLVDDD327 pKa = 4.72DD328 pKa = 4.45VCFRR332 pKa = 11.84EEE334 pKa = 4.78DD335 pKa = 3.55QCLVDDD341 pKa = 4.95DD342 pKa = 4.92VCFRR346 pKa = 11.84EEE348 pKa = 4.88DD349 pKa = 3.61QCLVDDD355 pKa = 3.71EE356 pKa = 5.03VCFRR360 pKa = 11.84EEE362 pKa = 5.01DD363 pKa = 3.64QCLVDDD369 pKa = 3.71EE370 pKa = 5.03VCFRR374 pKa = 11.84EEE376 pKa = 4.67DD377 pKa = 3.36QSLVDDD383 pKa = 3.22EE384 pKa = 4.56VRR386 pKa = 11.84FRR388 pKa = 11.84EEE390 pKa = 4.53DD391 pKa = 3.18QSLVDDD397 pKa = 4.34DD398 pKa = 4.72VCFRR402 pKa = 11.84EEE404 pKa = 4.28EE405 pKa = 4.17QSLVDDD411 pKa = 4.28DD412 pKa = 4.89VCFRR416 pKa = 11.84SRR418 pKa = 11.84QSVSG
HHH2 pKa = 7.77LYYY5 pKa = 11.04RR6 pKa = 11.84DDD8 pKa = 3.41KK9 pKa = 10.26QIKKK13 pKa = 7.51KKK15 pKa = 9.87QLGSVRR21 pKa = 11.84CHHH24 pKa = 7.43EEE26 pKa = 3.54DD27 pKa = 4.05DD28 pKa = 3.51CLVDDD33 pKa = 5.31DD34 pKa = 4.66VCFRR38 pKa = 11.84EEE40 pKa = 4.78DD41 pKa = 3.55QCLVDDD47 pKa = 4.72DD48 pKa = 4.45VCFRR52 pKa = 11.84EEE54 pKa = 4.46DD55 pKa = 3.26QSLVDDD61 pKa = 4.34DD62 pKa = 4.79VCFRR66 pKa = 11.84EEE68 pKa = 4.88DD69 pKa = 3.72QCLVDDD75 pKa = 4.72DD76 pKa = 4.45VCFRR80 pKa = 11.84EEE82 pKa = 4.46DD83 pKa = 3.17QSLVDDD89 pKa = 3.49EE90 pKa = 4.91VCFRR94 pKa = 11.84EEE96 pKa = 4.84DD97 pKa = 3.51QSLLDDD103 pKa = 3.55EE104 pKa = 4.59VCFRR108 pKa = 11.84EEE110 pKa = 4.67DD111 pKa = 3.36QSLVDDD117 pKa = 3.49EE118 pKa = 4.91VCFRR122 pKa = 11.84EEE124 pKa = 4.67DD125 pKa = 3.36QSLVDDD131 pKa = 3.49EE132 pKa = 4.91VCFRR136 pKa = 11.84EEE138 pKa = 4.67DD139 pKa = 3.36QSLVDDD145 pKa = 3.49EE146 pKa = 4.91VCFRR150 pKa = 11.84EEE152 pKa = 5.01DD153 pKa = 3.64QCLVDDD159 pKa = 3.71EE160 pKa = 5.03VCFRR164 pKa = 11.84EEE166 pKa = 5.01DD167 pKa = 3.73QCLVDDD173 pKa = 4.95DD174 pKa = 4.92VCFRR178 pKa = 11.84EEE180 pKa = 4.57DD181 pKa = 3.44QSLVDDD187 pKa = 5.05DD188 pKa = 4.6EE189 pKa = 4.78CLRR192 pKa = 11.84EEE194 pKa = 4.48GQCLVDDD201 pKa = 3.38EE202 pKa = 5.04VCFRR206 pKa = 11.84EEE208 pKa = 5.07GQSLVDDD215 pKa = 3.32EE216 pKa = 4.92VCFRR220 pKa = 11.84EEE222 pKa = 4.67DD223 pKa = 3.45QSLVDDD229 pKa = 4.34DD230 pKa = 4.79VCFRR234 pKa = 11.84EEE236 pKa = 4.57DD237 pKa = 3.44QSLVDDD243 pKa = 3.95DD244 pKa = 4.24VCFRR248 pKa = 11.84EEE250 pKa = 4.62GQCLVDDD257 pKa = 3.38EE258 pKa = 5.05VCFRR262 pKa = 11.84EEE264 pKa = 4.67DD265 pKa = 3.45QSLVDDD271 pKa = 4.34DD272 pKa = 4.79VCFRR276 pKa = 11.84EEE278 pKa = 4.88DD279 pKa = 3.72QCLVDDD285 pKa = 4.95DD286 pKa = 4.92VCFRR290 pKa = 11.84EEE292 pKa = 4.88DD293 pKa = 3.72QCLVDDD299 pKa = 4.95DD300 pKa = 4.92VCFRR304 pKa = 11.84EEE306 pKa = 4.88DD307 pKa = 3.72QCLVDDD313 pKa = 4.95DD314 pKa = 4.92VCFRR318 pKa = 11.84EEE320 pKa = 4.88DD321 pKa = 3.72QCLVDDD327 pKa = 4.72DD328 pKa = 4.45VCFRR332 pKa = 11.84EEE334 pKa = 4.78DD335 pKa = 3.55QCLVDDD341 pKa = 4.95DD342 pKa = 4.92VCFRR346 pKa = 11.84EEE348 pKa = 4.88DD349 pKa = 3.61QCLVDDD355 pKa = 3.71EE356 pKa = 5.03VCFRR360 pKa = 11.84EEE362 pKa = 5.01DD363 pKa = 3.64QCLVDDD369 pKa = 3.71EE370 pKa = 5.03VCFRR374 pKa = 11.84EEE376 pKa = 4.67DD377 pKa = 3.36QSLVDDD383 pKa = 3.22EE384 pKa = 4.56VRR386 pKa = 11.84FRR388 pKa = 11.84EEE390 pKa = 4.53DD391 pKa = 3.18QSLVDDD397 pKa = 4.34DD398 pKa = 4.72VCFRR402 pKa = 11.84EEE404 pKa = 4.28EE405 pKa = 4.17QSLVDDD411 pKa = 4.28DD412 pKa = 4.89VCFRR416 pKa = 11.84SRR418 pKa = 11.84QSVSG
Molecular weight: 47.13 kDa
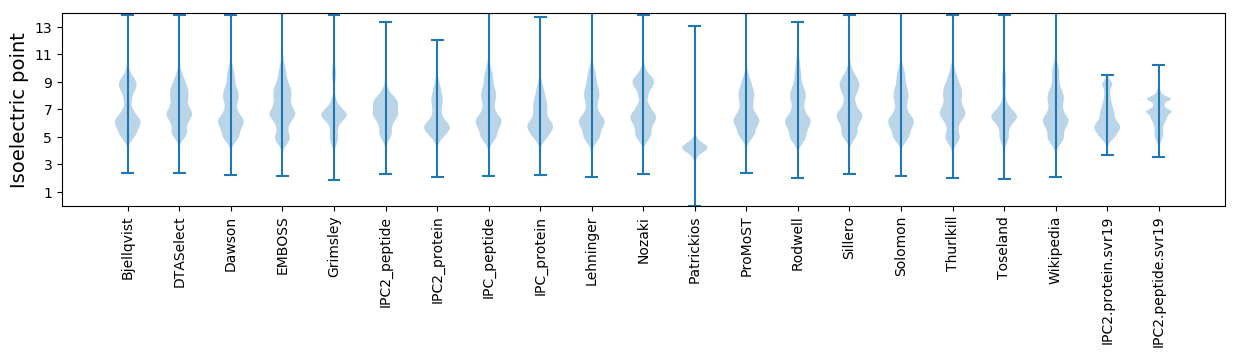
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6Q2ZIK4|A0A6Q2ZIK4_ESOLU NF-kappa-B inhibitor alpha OS=Esox lucius OX=8010 PE=4 SV=1
MM1 pKa = 7.2QGISVPINTPRR12 pKa = 11.84TRR14 pKa = 11.84VPINTPRR21 pKa = 11.84TRR23 pKa = 11.84VPINTPRR30 pKa = 11.84TRR32 pKa = 11.84VPINTPRR39 pKa = 11.84TRR41 pKa = 11.84VPINTPRR48 pKa = 11.84TRR50 pKa = 11.84VPINTPRR57 pKa = 11.84TRR59 pKa = 11.84VPINTPRR66 pKa = 11.84TRR68 pKa = 11.84VPINTPRR75 pKa = 11.84TRR77 pKa = 11.84VPINTPRR84 pKa = 11.84TRR86 pKa = 11.84VPINTPRR93 pKa = 11.84TRR95 pKa = 11.84VPINTPRR102 pKa = 11.84TRR104 pKa = 11.84VPINTPRR111 pKa = 11.84TRR113 pKa = 11.84VPINTPRR120 pKa = 11.84TRR122 pKa = 11.84SGVSAPTPNPGIDD135 pKa = 3.48QSSHH139 pKa = 6.38SII141 pKa = 3.44
MM1 pKa = 7.2QGISVPINTPRR12 pKa = 11.84TRR14 pKa = 11.84VPINTPRR21 pKa = 11.84TRR23 pKa = 11.84VPINTPRR30 pKa = 11.84TRR32 pKa = 11.84VPINTPRR39 pKa = 11.84TRR41 pKa = 11.84VPINTPRR48 pKa = 11.84TRR50 pKa = 11.84VPINTPRR57 pKa = 11.84TRR59 pKa = 11.84VPINTPRR66 pKa = 11.84TRR68 pKa = 11.84VPINTPRR75 pKa = 11.84TRR77 pKa = 11.84VPINTPRR84 pKa = 11.84TRR86 pKa = 11.84VPINTPRR93 pKa = 11.84TRR95 pKa = 11.84VPINTPRR102 pKa = 11.84TRR104 pKa = 11.84VPINTPRR111 pKa = 11.84TRR113 pKa = 11.84VPINTPRR120 pKa = 11.84TRR122 pKa = 11.84SGVSAPTPNPGIDD135 pKa = 3.48QSSHH139 pKa = 6.38SII141 pKa = 3.44
Molecular weight: 15.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
50391987 |
18 |
8678 |
704.6 |
78.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.342 ± 0.007 | 2.322 ± 0.007 |
5.233 ± 0.006 | 6.705 ± 0.011 |
3.714 ± 0.006 | 6.555 ± 0.008 |
2.667 ± 0.004 | 4.51 ± 0.006 |
5.495 ± 0.009 | 9.673 ± 0.01 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.432 ± 0.003 | 3.903 ± 0.005 |
5.672 ± 0.01 | 4.605 ± 0.007 |
5.625 ± 0.006 | 8.212 ± 0.009 |
5.707 ± 0.01 | 6.555 ± 0.007 |
1.197 ± 0.003 | 2.876 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
