
Sphingobium sp. YR768
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingobium; unclassified Sphingobium
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
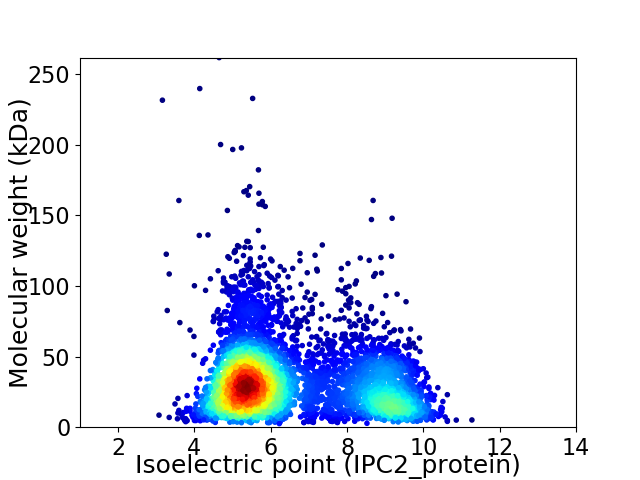
Virtual 2D-PAGE plot for 5343 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
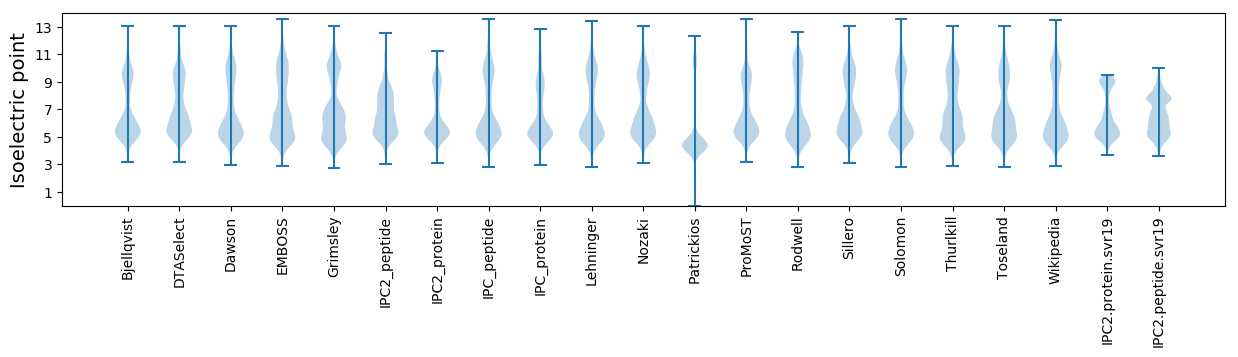
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9U428|A0A1H9U428_9SPHN Lipopolysaccharide export system protein LptA OS=Sphingobium sp. YR768 OX=1884365 GN=SAMN05518866_13326 PE=4 SV=1
MM1 pKa = 7.41GEE3 pKa = 4.0SVAIFDD9 pKa = 5.3GGLEE13 pKa = 3.95DD14 pKa = 6.82DD15 pKa = 4.78IYY17 pKa = 11.36LGTKK21 pKa = 10.01SSDD24 pKa = 3.28IISGNGGNDD33 pKa = 3.17ALVGDD38 pKa = 4.75EE39 pKa = 5.88GSDD42 pKa = 3.94QIDD45 pKa = 3.66GGDD48 pKa = 3.85GNDD51 pKa = 3.44FLYY54 pKa = 10.85AQTLSFDD61 pKa = 2.98WKK63 pKa = 10.5GYY65 pKa = 6.88NFRR68 pKa = 11.84WLPPVTDD75 pKa = 3.21VDD77 pKa = 4.19AVKK80 pKa = 9.71DD81 pKa = 3.85TLNGGDD87 pKa = 3.95GNDD90 pKa = 4.79HH91 pKa = 6.72ISAGYY96 pKa = 10.44GDD98 pKa = 5.86DD99 pKa = 3.45IDD101 pKa = 5.05GGEE104 pKa = 4.16GSDD107 pKa = 3.51YY108 pKa = 11.19LVISFRR114 pKa = 11.84GASSGIIADD123 pKa = 4.66FSTLEE128 pKa = 4.03TGGSVTVEE136 pKa = 3.59GGTIKK141 pKa = 10.52NVEE144 pKa = 3.99MLEE147 pKa = 4.2WVDD150 pKa = 3.66GSEE153 pKa = 4.19FSDD156 pKa = 4.3VITGQIAASDD166 pKa = 3.77APIYY170 pKa = 10.78GLGGDD175 pKa = 3.97DD176 pKa = 4.17HH177 pKa = 7.79LIAGGTGTGRR187 pKa = 11.84LYY189 pKa = 11.14GGDD192 pKa = 3.54GDD194 pKa = 4.8DD195 pKa = 4.03TLEE198 pKa = 4.48SAWSMLYY205 pKa = 10.6GGSGNDD211 pKa = 3.98LLIGGYY217 pKa = 9.28PFAYY221 pKa = 10.26GEE223 pKa = 5.0DD224 pKa = 3.9GDD226 pKa = 4.56DD227 pKa = 5.03VIWALNEE234 pKa = 3.86ASGGAGDD241 pKa = 5.63DD242 pKa = 3.36IIGFLRR248 pKa = 11.84DD249 pKa = 3.2LDD251 pKa = 3.93GSGRR255 pKa = 11.84PGILYY260 pKa = 10.54GDD262 pKa = 4.8DD263 pKa = 3.61GNDD266 pKa = 3.18RR267 pKa = 11.84LEE269 pKa = 4.53GSGHH273 pKa = 6.1IDD275 pKa = 3.21KK276 pKa = 11.11LSGGSGSDD284 pKa = 2.79ILYY287 pKa = 10.94GNGGDD292 pKa = 4.43DD293 pKa = 3.35ILYY296 pKa = 10.54AGASGSGTGAFDD308 pKa = 4.37PSDD311 pKa = 3.49SLAEE315 pKa = 4.07HH316 pKa = 6.89DD317 pKa = 4.46QLFGGDD323 pKa = 4.18GDD325 pKa = 4.14DD326 pKa = 4.65VMIIGYY332 pKa = 10.43GDD334 pKa = 3.91DD335 pKa = 4.09ADD337 pKa = 5.17GGDD340 pKa = 3.86GTDD343 pKa = 3.1SVTVYY348 pKa = 11.44ALGASSGVFIDD359 pKa = 5.34IGDD362 pKa = 4.01PSNGRR367 pKa = 11.84IGTGIIRR374 pKa = 11.84NVEE377 pKa = 4.0IIIGVYY383 pKa = 10.51ASDD386 pKa = 3.8FDD388 pKa = 5.58DD389 pKa = 4.6VIKK392 pKa = 10.8VDD394 pKa = 3.94NVGSDD399 pKa = 3.18ASINGKK405 pKa = 9.57KK406 pKa = 10.72GNDD409 pKa = 3.46IISGLSHH416 pKa = 6.65SVTFNGNEE424 pKa = 4.08GNDD427 pKa = 3.48ILIGSAQGDD436 pKa = 4.01MLDD439 pKa = 4.27GGSGNDD445 pKa = 3.4IIYY448 pKa = 10.61GGGGIDD454 pKa = 4.56RR455 pKa = 11.84ISGGDD460 pKa = 3.4GDD462 pKa = 5.37DD463 pKa = 3.99VIEE466 pKa = 4.96GGSGTNYY473 pKa = 9.68IGGGAGIDD481 pKa = 3.77TISYY485 pKa = 8.8RR486 pKa = 11.84HH487 pKa = 5.14STGAVVVDD495 pKa = 4.24LTPPDD500 pKa = 3.66GATVKK505 pKa = 10.54VQTPDD510 pKa = 3.4SIDD513 pKa = 3.34YY514 pKa = 10.4FSTIEE519 pKa = 4.07NVVGSQYY526 pKa = 11.32GDD528 pKa = 3.76SIKK531 pKa = 10.72GDD533 pKa = 3.27SGINVISGEE542 pKa = 4.46DD543 pKa = 3.73GDD545 pKa = 4.76DD546 pKa = 3.95TILGQDD552 pKa = 3.93GADD555 pKa = 3.3ILYY558 pKa = 10.59GDD560 pKa = 5.03AGNDD564 pKa = 3.42VLGGGVGIDD573 pKa = 3.38TLYY576 pKa = 11.33GGTGNDD582 pKa = 3.81RR583 pKa = 11.84LDD585 pKa = 3.91GGAGADD591 pKa = 3.42RR592 pKa = 11.84LVGGVGDD599 pKa = 5.4DD600 pKa = 3.31IYY602 pKa = 10.98IVDD605 pKa = 3.74TAGDD609 pKa = 4.15VIVEE613 pKa = 4.13LAGEE617 pKa = 4.58GDD619 pKa = 3.62DD620 pKa = 4.35HH621 pKa = 7.9VYY623 pKa = 11.17SSVSYY628 pKa = 10.24ILSSHH633 pKa = 6.27IEE635 pKa = 4.08RR636 pKa = 11.84LSMTGTAAVDD646 pKa = 3.7GTGNEE651 pKa = 4.13LDD653 pKa = 3.67NVIVGNAAANKK664 pKa = 8.39LTGGTGNDD672 pKa = 3.73YY673 pKa = 11.35LDD675 pKa = 4.71GLDD678 pKa = 5.28GDD680 pKa = 5.09DD681 pKa = 4.7LLEE684 pKa = 4.83GGAGNDD690 pKa = 3.13ILIGGRR696 pKa = 11.84GGDD699 pKa = 3.62QMIGGAGDD707 pKa = 3.82DD708 pKa = 4.57LYY710 pKa = 11.76VVDD713 pKa = 6.27DD714 pKa = 3.76IRR716 pKa = 11.84DD717 pKa = 3.99TVSEE721 pKa = 4.43DD722 pKa = 2.96ADD724 pKa = 3.69AGIDD728 pKa = 3.68TVQSSVSFVLGDD740 pKa = 3.6NVEE743 pKa = 3.97NLALGGTAALTGTGNALNNVMTGNSGVSTLLGGDD777 pKa = 3.83GDD779 pKa = 4.37DD780 pKa = 4.38HH781 pKa = 7.45LSSGVNGRR789 pKa = 11.84GILVGGAGSDD799 pKa = 3.69VLIGSAGNDD808 pKa = 3.45SLNSGLSGVRR818 pKa = 11.84DD819 pKa = 3.57MGTEE823 pKa = 3.79KK824 pKa = 10.92DD825 pKa = 3.78VLIAGAGDD833 pKa = 4.15DD834 pKa = 4.39GASAGYY840 pKa = 10.58GDD842 pKa = 5.51DD843 pKa = 3.54VDD845 pKa = 4.83GGEE848 pKa = 5.12GYY850 pKa = 10.82DD851 pKa = 3.44SLSYY855 pKa = 11.22SFGGASAGIDD865 pKa = 3.2IRR867 pKa = 11.84SSDD870 pKa = 3.92LLGAGPHH877 pKa = 5.96VIGGGTVQNFEE888 pKa = 4.45RR889 pKa = 11.84LSGLRR894 pKa = 11.84GSEE897 pKa = 3.48FDD899 pKa = 5.57DD900 pKa = 4.18YY901 pKa = 11.69INYY904 pKa = 7.76DD905 pKa = 3.44TFYY908 pKa = 10.54ISQLVIEE915 pKa = 5.14AGAGNDD921 pKa = 3.62TVIITGDD928 pKa = 3.76APVKK932 pKa = 10.18MLGGDD937 pKa = 3.42GDD939 pKa = 4.02DD940 pKa = 3.74RR941 pKa = 11.84LVVGYY946 pKa = 10.08QPGNQFDD953 pKa = 4.12GGAGSDD959 pKa = 3.56TIDD962 pKa = 3.11FRR964 pKa = 11.84LYY966 pKa = 10.43GEE968 pKa = 4.52KK969 pKa = 9.58VTVALGLNGANGNGGPYY986 pKa = 9.0LTQLLNVEE994 pKa = 4.29NVIGSSYY1001 pKa = 11.35ADD1003 pKa = 3.57GIVGNEE1009 pKa = 4.0LANDD1013 pKa = 3.67LRR1015 pKa = 11.84GGDD1018 pKa = 3.96GDD1020 pKa = 4.45DD1021 pKa = 4.56TILGQDD1027 pKa = 4.17GADD1030 pKa = 3.69KK1031 pKa = 11.06LYY1033 pKa = 11.18GDD1035 pKa = 5.13AGNDD1039 pKa = 3.51VLGGGAGIDD1048 pKa = 3.5TLYY1051 pKa = 11.22GGTGNDD1057 pKa = 3.81RR1058 pKa = 11.84LDD1060 pKa = 3.91GGAGADD1066 pKa = 3.42RR1067 pKa = 11.84LVGGVGDD1074 pKa = 5.4DD1075 pKa = 3.31IYY1077 pKa = 10.98IVDD1080 pKa = 3.74TAGDD1084 pKa = 4.15VIVEE1088 pKa = 4.12LAGEE1092 pKa = 4.44GNDD1095 pKa = 3.37TVVASGSYY1103 pKa = 8.88TLSDD1107 pKa = 3.19HH1108 pKa = 7.62VEE1110 pKa = 4.08TLKK1113 pKa = 11.43LNGTVGAADD1122 pKa = 3.54MRR1124 pKa = 11.84GNGQDD1129 pKa = 3.53NLITAEE1135 pKa = 4.19GASGSSALGIRR1146 pKa = 11.84LFGLGGNDD1154 pKa = 3.15MLLGSVAGDD1163 pKa = 3.49VLDD1166 pKa = 4.77GGTGNDD1172 pKa = 3.32VMKK1175 pKa = 10.87GGLGSDD1181 pKa = 4.26LYY1183 pKa = 11.47YY1184 pKa = 10.58VDD1186 pKa = 4.8SVGDD1190 pKa = 3.66QIVEE1194 pKa = 4.08GLNAGVDD1201 pKa = 3.97HH1202 pKa = 6.38VASSISYY1209 pKa = 8.18TLGANVEE1216 pKa = 4.34GLTLTGSANLNGIGNALSNAIFGNDD1241 pKa = 3.1GNNYY1245 pKa = 9.11LSGMGGRR1252 pKa = 11.84DD1253 pKa = 3.08TLYY1256 pKa = 10.66GYY1258 pKa = 10.49GGNDD1262 pKa = 3.08VLVPGDD1268 pKa = 3.74GNNVVDD1274 pKa = 4.68GGVGYY1279 pKa = 10.57DD1280 pKa = 3.79LLLLTGAKK1288 pKa = 9.94GSYY1291 pKa = 10.11SYY1293 pKa = 11.45LSANGSIYY1301 pKa = 10.72LVGEE1305 pKa = 4.1EE1306 pKa = 5.07GATKK1310 pKa = 7.95MTGVEE1315 pKa = 4.26SVRR1318 pKa = 11.84FADD1321 pKa = 3.95GTVATGDD1328 pKa = 3.8LASSLTAFDD1337 pKa = 3.58GLRR1340 pKa = 11.84YY1341 pKa = 9.01IAGYY1345 pKa = 9.26SDD1347 pKa = 3.5LMAVFGTNAAAGVSHH1362 pKa = 5.95YY1363 pKa = 10.51VSSGFAEE1370 pKa = 4.4GRR1372 pKa = 11.84DD1373 pKa = 3.33AKK1375 pKa = 11.21SFDD1378 pKa = 3.81PLDD1381 pKa = 4.21YY1382 pKa = 10.7IAGYY1386 pKa = 10.29SDD1388 pKa = 3.8LASAFGTDD1396 pKa = 2.96AQAATRR1402 pKa = 11.84HH1403 pKa = 5.84YY1404 pKa = 8.95ITTGAKK1410 pKa = 9.68EE1411 pKa = 3.9GRR1413 pKa = 11.84NDD1415 pKa = 4.22DD1416 pKa = 4.17RR1417 pKa = 11.84FDD1419 pKa = 3.97ALDD1422 pKa = 3.66YY1423 pKa = 10.39LASYY1427 pKa = 10.9ADD1429 pKa = 4.11LSATLGADD1437 pKa = 3.37EE1438 pKa = 4.33EE1439 pKa = 4.73AGARR1443 pKa = 11.84HH1444 pKa = 6.15YY1445 pKa = 10.2LTIGKK1450 pKa = 8.89AQGRR1454 pKa = 11.84SDD1456 pKa = 4.38DD1457 pKa = 4.37GFDD1460 pKa = 3.41GLQYY1464 pKa = 11.0AAGYY1468 pKa = 10.43ADD1470 pKa = 4.43LAALFGSNDD1479 pKa = 3.42DD1480 pKa = 3.48AAARR1484 pKa = 11.84HH1485 pKa = 5.44YY1486 pKa = 10.08IQYY1489 pKa = 10.07GRR1491 pKa = 11.84AEE1493 pKa = 4.09GRR1495 pKa = 11.84VADD1498 pKa = 4.22KK1499 pKa = 10.93FDD1501 pKa = 3.29GLRR1504 pKa = 11.84YY1505 pKa = 9.19IASNADD1511 pKa = 4.13LIVSIGDD1518 pKa = 3.52NDD1520 pKa = 3.82DD1521 pKa = 3.87AAAKK1525 pKa = 10.04HH1526 pKa = 5.55YY1527 pKa = 10.05LDD1529 pKa = 4.77HH1530 pKa = 6.98GFAEE1534 pKa = 4.67HH1535 pKa = 7.57RR1536 pKa = 11.84STTSFDD1542 pKa = 3.42ALKK1545 pKa = 10.52YY1546 pKa = 10.17AAANPDD1552 pKa = 3.06LAAAFGSDD1560 pKa = 3.33VEE1562 pKa = 4.81ALTEE1566 pKa = 4.01HH1567 pKa = 6.67YY1568 pKa = 10.34IDD1570 pKa = 3.8FGYY1573 pKa = 10.74YY1574 pKa = 6.32EE1575 pKa = 4.5HH1576 pKa = 7.73RR1577 pKa = 11.84AIAPAAAMVEE1587 pKa = 4.61VIGG1590 pKa = 4.83
MM1 pKa = 7.41GEE3 pKa = 4.0SVAIFDD9 pKa = 5.3GGLEE13 pKa = 3.95DD14 pKa = 6.82DD15 pKa = 4.78IYY17 pKa = 11.36LGTKK21 pKa = 10.01SSDD24 pKa = 3.28IISGNGGNDD33 pKa = 3.17ALVGDD38 pKa = 4.75EE39 pKa = 5.88GSDD42 pKa = 3.94QIDD45 pKa = 3.66GGDD48 pKa = 3.85GNDD51 pKa = 3.44FLYY54 pKa = 10.85AQTLSFDD61 pKa = 2.98WKK63 pKa = 10.5GYY65 pKa = 6.88NFRR68 pKa = 11.84WLPPVTDD75 pKa = 3.21VDD77 pKa = 4.19AVKK80 pKa = 9.71DD81 pKa = 3.85TLNGGDD87 pKa = 3.95GNDD90 pKa = 4.79HH91 pKa = 6.72ISAGYY96 pKa = 10.44GDD98 pKa = 5.86DD99 pKa = 3.45IDD101 pKa = 5.05GGEE104 pKa = 4.16GSDD107 pKa = 3.51YY108 pKa = 11.19LVISFRR114 pKa = 11.84GASSGIIADD123 pKa = 4.66FSTLEE128 pKa = 4.03TGGSVTVEE136 pKa = 3.59GGTIKK141 pKa = 10.52NVEE144 pKa = 3.99MLEE147 pKa = 4.2WVDD150 pKa = 3.66GSEE153 pKa = 4.19FSDD156 pKa = 4.3VITGQIAASDD166 pKa = 3.77APIYY170 pKa = 10.78GLGGDD175 pKa = 3.97DD176 pKa = 4.17HH177 pKa = 7.79LIAGGTGTGRR187 pKa = 11.84LYY189 pKa = 11.14GGDD192 pKa = 3.54GDD194 pKa = 4.8DD195 pKa = 4.03TLEE198 pKa = 4.48SAWSMLYY205 pKa = 10.6GGSGNDD211 pKa = 3.98LLIGGYY217 pKa = 9.28PFAYY221 pKa = 10.26GEE223 pKa = 5.0DD224 pKa = 3.9GDD226 pKa = 4.56DD227 pKa = 5.03VIWALNEE234 pKa = 3.86ASGGAGDD241 pKa = 5.63DD242 pKa = 3.36IIGFLRR248 pKa = 11.84DD249 pKa = 3.2LDD251 pKa = 3.93GSGRR255 pKa = 11.84PGILYY260 pKa = 10.54GDD262 pKa = 4.8DD263 pKa = 3.61GNDD266 pKa = 3.18RR267 pKa = 11.84LEE269 pKa = 4.53GSGHH273 pKa = 6.1IDD275 pKa = 3.21KK276 pKa = 11.11LSGGSGSDD284 pKa = 2.79ILYY287 pKa = 10.94GNGGDD292 pKa = 4.43DD293 pKa = 3.35ILYY296 pKa = 10.54AGASGSGTGAFDD308 pKa = 4.37PSDD311 pKa = 3.49SLAEE315 pKa = 4.07HH316 pKa = 6.89DD317 pKa = 4.46QLFGGDD323 pKa = 4.18GDD325 pKa = 4.14DD326 pKa = 4.65VMIIGYY332 pKa = 10.43GDD334 pKa = 3.91DD335 pKa = 4.09ADD337 pKa = 5.17GGDD340 pKa = 3.86GTDD343 pKa = 3.1SVTVYY348 pKa = 11.44ALGASSGVFIDD359 pKa = 5.34IGDD362 pKa = 4.01PSNGRR367 pKa = 11.84IGTGIIRR374 pKa = 11.84NVEE377 pKa = 4.0IIIGVYY383 pKa = 10.51ASDD386 pKa = 3.8FDD388 pKa = 5.58DD389 pKa = 4.6VIKK392 pKa = 10.8VDD394 pKa = 3.94NVGSDD399 pKa = 3.18ASINGKK405 pKa = 9.57KK406 pKa = 10.72GNDD409 pKa = 3.46IISGLSHH416 pKa = 6.65SVTFNGNEE424 pKa = 4.08GNDD427 pKa = 3.48ILIGSAQGDD436 pKa = 4.01MLDD439 pKa = 4.27GGSGNDD445 pKa = 3.4IIYY448 pKa = 10.61GGGGIDD454 pKa = 4.56RR455 pKa = 11.84ISGGDD460 pKa = 3.4GDD462 pKa = 5.37DD463 pKa = 3.99VIEE466 pKa = 4.96GGSGTNYY473 pKa = 9.68IGGGAGIDD481 pKa = 3.77TISYY485 pKa = 8.8RR486 pKa = 11.84HH487 pKa = 5.14STGAVVVDD495 pKa = 4.24LTPPDD500 pKa = 3.66GATVKK505 pKa = 10.54VQTPDD510 pKa = 3.4SIDD513 pKa = 3.34YY514 pKa = 10.4FSTIEE519 pKa = 4.07NVVGSQYY526 pKa = 11.32GDD528 pKa = 3.76SIKK531 pKa = 10.72GDD533 pKa = 3.27SGINVISGEE542 pKa = 4.46DD543 pKa = 3.73GDD545 pKa = 4.76DD546 pKa = 3.95TILGQDD552 pKa = 3.93GADD555 pKa = 3.3ILYY558 pKa = 10.59GDD560 pKa = 5.03AGNDD564 pKa = 3.42VLGGGVGIDD573 pKa = 3.38TLYY576 pKa = 11.33GGTGNDD582 pKa = 3.81RR583 pKa = 11.84LDD585 pKa = 3.91GGAGADD591 pKa = 3.42RR592 pKa = 11.84LVGGVGDD599 pKa = 5.4DD600 pKa = 3.31IYY602 pKa = 10.98IVDD605 pKa = 3.74TAGDD609 pKa = 4.15VIVEE613 pKa = 4.13LAGEE617 pKa = 4.58GDD619 pKa = 3.62DD620 pKa = 4.35HH621 pKa = 7.9VYY623 pKa = 11.17SSVSYY628 pKa = 10.24ILSSHH633 pKa = 6.27IEE635 pKa = 4.08RR636 pKa = 11.84LSMTGTAAVDD646 pKa = 3.7GTGNEE651 pKa = 4.13LDD653 pKa = 3.67NVIVGNAAANKK664 pKa = 8.39LTGGTGNDD672 pKa = 3.73YY673 pKa = 11.35LDD675 pKa = 4.71GLDD678 pKa = 5.28GDD680 pKa = 5.09DD681 pKa = 4.7LLEE684 pKa = 4.83GGAGNDD690 pKa = 3.13ILIGGRR696 pKa = 11.84GGDD699 pKa = 3.62QMIGGAGDD707 pKa = 3.82DD708 pKa = 4.57LYY710 pKa = 11.76VVDD713 pKa = 6.27DD714 pKa = 3.76IRR716 pKa = 11.84DD717 pKa = 3.99TVSEE721 pKa = 4.43DD722 pKa = 2.96ADD724 pKa = 3.69AGIDD728 pKa = 3.68TVQSSVSFVLGDD740 pKa = 3.6NVEE743 pKa = 3.97NLALGGTAALTGTGNALNNVMTGNSGVSTLLGGDD777 pKa = 3.83GDD779 pKa = 4.37DD780 pKa = 4.38HH781 pKa = 7.45LSSGVNGRR789 pKa = 11.84GILVGGAGSDD799 pKa = 3.69VLIGSAGNDD808 pKa = 3.45SLNSGLSGVRR818 pKa = 11.84DD819 pKa = 3.57MGTEE823 pKa = 3.79KK824 pKa = 10.92DD825 pKa = 3.78VLIAGAGDD833 pKa = 4.15DD834 pKa = 4.39GASAGYY840 pKa = 10.58GDD842 pKa = 5.51DD843 pKa = 3.54VDD845 pKa = 4.83GGEE848 pKa = 5.12GYY850 pKa = 10.82DD851 pKa = 3.44SLSYY855 pKa = 11.22SFGGASAGIDD865 pKa = 3.2IRR867 pKa = 11.84SSDD870 pKa = 3.92LLGAGPHH877 pKa = 5.96VIGGGTVQNFEE888 pKa = 4.45RR889 pKa = 11.84LSGLRR894 pKa = 11.84GSEE897 pKa = 3.48FDD899 pKa = 5.57DD900 pKa = 4.18YY901 pKa = 11.69INYY904 pKa = 7.76DD905 pKa = 3.44TFYY908 pKa = 10.54ISQLVIEE915 pKa = 5.14AGAGNDD921 pKa = 3.62TVIITGDD928 pKa = 3.76APVKK932 pKa = 10.18MLGGDD937 pKa = 3.42GDD939 pKa = 4.02DD940 pKa = 3.74RR941 pKa = 11.84LVVGYY946 pKa = 10.08QPGNQFDD953 pKa = 4.12GGAGSDD959 pKa = 3.56TIDD962 pKa = 3.11FRR964 pKa = 11.84LYY966 pKa = 10.43GEE968 pKa = 4.52KK969 pKa = 9.58VTVALGLNGANGNGGPYY986 pKa = 9.0LTQLLNVEE994 pKa = 4.29NVIGSSYY1001 pKa = 11.35ADD1003 pKa = 3.57GIVGNEE1009 pKa = 4.0LANDD1013 pKa = 3.67LRR1015 pKa = 11.84GGDD1018 pKa = 3.96GDD1020 pKa = 4.45DD1021 pKa = 4.56TILGQDD1027 pKa = 4.17GADD1030 pKa = 3.69KK1031 pKa = 11.06LYY1033 pKa = 11.18GDD1035 pKa = 5.13AGNDD1039 pKa = 3.51VLGGGAGIDD1048 pKa = 3.5TLYY1051 pKa = 11.22GGTGNDD1057 pKa = 3.81RR1058 pKa = 11.84LDD1060 pKa = 3.91GGAGADD1066 pKa = 3.42RR1067 pKa = 11.84LVGGVGDD1074 pKa = 5.4DD1075 pKa = 3.31IYY1077 pKa = 10.98IVDD1080 pKa = 3.74TAGDD1084 pKa = 4.15VIVEE1088 pKa = 4.12LAGEE1092 pKa = 4.44GNDD1095 pKa = 3.37TVVASGSYY1103 pKa = 8.88TLSDD1107 pKa = 3.19HH1108 pKa = 7.62VEE1110 pKa = 4.08TLKK1113 pKa = 11.43LNGTVGAADD1122 pKa = 3.54MRR1124 pKa = 11.84GNGQDD1129 pKa = 3.53NLITAEE1135 pKa = 4.19GASGSSALGIRR1146 pKa = 11.84LFGLGGNDD1154 pKa = 3.15MLLGSVAGDD1163 pKa = 3.49VLDD1166 pKa = 4.77GGTGNDD1172 pKa = 3.32VMKK1175 pKa = 10.87GGLGSDD1181 pKa = 4.26LYY1183 pKa = 11.47YY1184 pKa = 10.58VDD1186 pKa = 4.8SVGDD1190 pKa = 3.66QIVEE1194 pKa = 4.08GLNAGVDD1201 pKa = 3.97HH1202 pKa = 6.38VASSISYY1209 pKa = 8.18TLGANVEE1216 pKa = 4.34GLTLTGSANLNGIGNALSNAIFGNDD1241 pKa = 3.1GNNYY1245 pKa = 9.11LSGMGGRR1252 pKa = 11.84DD1253 pKa = 3.08TLYY1256 pKa = 10.66GYY1258 pKa = 10.49GGNDD1262 pKa = 3.08VLVPGDD1268 pKa = 3.74GNNVVDD1274 pKa = 4.68GGVGYY1279 pKa = 10.57DD1280 pKa = 3.79LLLLTGAKK1288 pKa = 9.94GSYY1291 pKa = 10.11SYY1293 pKa = 11.45LSANGSIYY1301 pKa = 10.72LVGEE1305 pKa = 4.1EE1306 pKa = 5.07GATKK1310 pKa = 7.95MTGVEE1315 pKa = 4.26SVRR1318 pKa = 11.84FADD1321 pKa = 3.95GTVATGDD1328 pKa = 3.8LASSLTAFDD1337 pKa = 3.58GLRR1340 pKa = 11.84YY1341 pKa = 9.01IAGYY1345 pKa = 9.26SDD1347 pKa = 3.5LMAVFGTNAAAGVSHH1362 pKa = 5.95YY1363 pKa = 10.51VSSGFAEE1370 pKa = 4.4GRR1372 pKa = 11.84DD1373 pKa = 3.33AKK1375 pKa = 11.21SFDD1378 pKa = 3.81PLDD1381 pKa = 4.21YY1382 pKa = 10.7IAGYY1386 pKa = 10.29SDD1388 pKa = 3.8LASAFGTDD1396 pKa = 2.96AQAATRR1402 pKa = 11.84HH1403 pKa = 5.84YY1404 pKa = 8.95ITTGAKK1410 pKa = 9.68EE1411 pKa = 3.9GRR1413 pKa = 11.84NDD1415 pKa = 4.22DD1416 pKa = 4.17RR1417 pKa = 11.84FDD1419 pKa = 3.97ALDD1422 pKa = 3.66YY1423 pKa = 10.39LASYY1427 pKa = 10.9ADD1429 pKa = 4.11LSATLGADD1437 pKa = 3.37EE1438 pKa = 4.33EE1439 pKa = 4.73AGARR1443 pKa = 11.84HH1444 pKa = 6.15YY1445 pKa = 10.2LTIGKK1450 pKa = 8.89AQGRR1454 pKa = 11.84SDD1456 pKa = 4.38DD1457 pKa = 4.37GFDD1460 pKa = 3.41GLQYY1464 pKa = 11.0AAGYY1468 pKa = 10.43ADD1470 pKa = 4.43LAALFGSNDD1479 pKa = 3.42DD1480 pKa = 3.48AAARR1484 pKa = 11.84HH1485 pKa = 5.44YY1486 pKa = 10.08IQYY1489 pKa = 10.07GRR1491 pKa = 11.84AEE1493 pKa = 4.09GRR1495 pKa = 11.84VADD1498 pKa = 4.22KK1499 pKa = 10.93FDD1501 pKa = 3.29GLRR1504 pKa = 11.84YY1505 pKa = 9.19IASNADD1511 pKa = 4.13LIVSIGDD1518 pKa = 3.52NDD1520 pKa = 3.82DD1521 pKa = 3.87AAAKK1525 pKa = 10.04HH1526 pKa = 5.55YY1527 pKa = 10.05LDD1529 pKa = 4.77HH1530 pKa = 6.98GFAEE1534 pKa = 4.67HH1535 pKa = 7.57RR1536 pKa = 11.84STTSFDD1542 pKa = 3.42ALKK1545 pKa = 10.52YY1546 pKa = 10.17AAANPDD1552 pKa = 3.06LAAAFGSDD1560 pKa = 3.33VEE1562 pKa = 4.81ALTEE1566 pKa = 4.01HH1567 pKa = 6.67YY1568 pKa = 10.34IDD1570 pKa = 3.8FGYY1573 pKa = 10.74YY1574 pKa = 6.32EE1575 pKa = 4.5HH1576 pKa = 7.73RR1577 pKa = 11.84AIAPAAAMVEE1587 pKa = 4.61VIGG1590 pKa = 4.83
Molecular weight: 160.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9KWK9|A0A1H9KWK9_9SPHN Proline iminopeptidase OS=Sphingobium sp. YR768 OX=1884365 GN=SAMN05518866_104189 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.06SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.06SLSAA44 pKa = 3.93
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1701863 |
24 |
2462 |
318.5 |
34.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.935 ± 0.05 | 0.794 ± 0.011 |
6.243 ± 0.025 | 5.017 ± 0.033 |
3.523 ± 0.024 | 8.845 ± 0.033 |
2.075 ± 0.019 | 5.239 ± 0.021 |
3.107 ± 0.027 | 9.803 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.545 ± 0.018 | 2.656 ± 0.023 |
5.281 ± 0.026 | 3.338 ± 0.018 |
7.143 ± 0.034 | 5.448 ± 0.026 |
5.287 ± 0.031 | 6.858 ± 0.025 |
1.472 ± 0.015 | 2.393 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
