
Phaseolus angularis (Azuki bean) (Vigna angularis)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; 50 kb inversion clade;
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
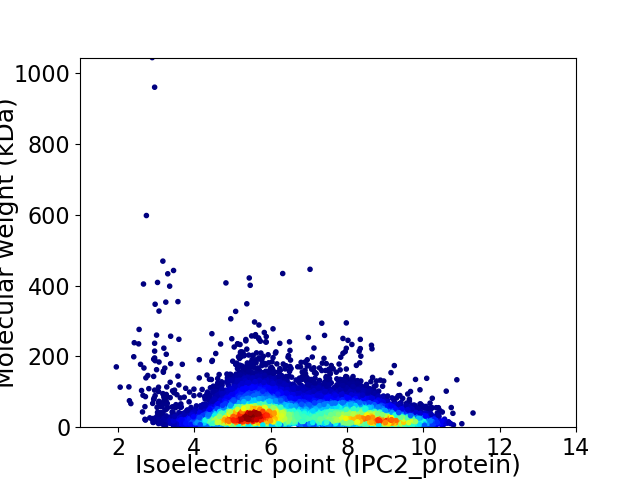
Virtual 2D-PAGE plot for 33730 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0L9TXU1|A0A0L9TXU1_PHAAN AP2/ERF domain-containing protein OS=Phaseolus angularis OX=3914 GN=LR48_Vigan02g136700 PE=4 SV=1
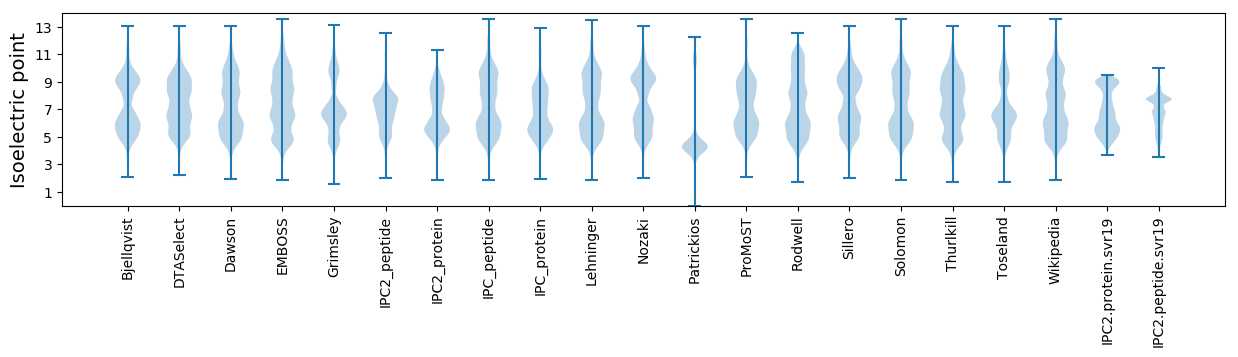
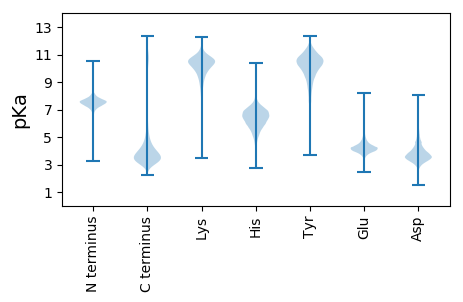
MM1 pKa = 7.32EE2 pKa = 5.44RR3 pKa = 11.84KK4 pKa = 9.66KK5 pKa = 10.64PLDD8 pKa = 3.61FSSYY12 pKa = 10.55FCFEE16 pKa = 3.89ASGDD20 pKa = 4.0SEE22 pKa = 4.37EE23 pKa = 4.7AHH25 pKa = 6.9PDD27 pKa = 3.49DD28 pKa = 6.21PIFACEE34 pKa = 3.7MTRR37 pKa = 11.84AYY39 pKa = 10.54GDD41 pKa = 4.44DD42 pKa = 4.01EE43 pKa = 6.11NDD45 pKa = 4.32DD46 pKa = 4.25ALSCNFEE53 pKa = 3.93GSGAFDD59 pKa = 3.03GAVFDD64 pKa = 4.45EE65 pKa = 5.32EE66 pKa = 5.16EE67 pKa = 4.47NEE69 pKa = 4.62CCDD72 pKa = 5.17DD73 pKa = 4.35DD74 pKa = 6.63HH75 pKa = 9.25DD76 pKa = 5.69DD77 pKa = 4.82EE78 pKa = 4.64IAKK81 pKa = 8.08VTQDD85 pKa = 3.63EE86 pKa = 4.43KK87 pKa = 11.04SGVYY91 pKa = 10.3GMSYY95 pKa = 10.89CEE97 pKa = 4.5DD98 pKa = 3.9DD99 pKa = 4.92DD100 pKa = 4.55MEE102 pKa = 4.39EE103 pKa = 4.04EE104 pKa = 4.3HH105 pKa = 6.65MKK107 pKa = 10.47SHH109 pKa = 6.27VSFDD113 pKa = 3.62SGHH116 pKa = 5.85EE117 pKa = 4.1FVDD120 pKa = 3.71EE121 pKa = 4.16MEE123 pKa = 4.39KK124 pKa = 10.71NRR126 pKa = 11.84LFWEE130 pKa = 4.55ACLASS135 pKa = 3.65
MM1 pKa = 7.32EE2 pKa = 5.44RR3 pKa = 11.84KK4 pKa = 9.66KK5 pKa = 10.64PLDD8 pKa = 3.61FSSYY12 pKa = 10.55FCFEE16 pKa = 3.89ASGDD20 pKa = 4.0SEE22 pKa = 4.37EE23 pKa = 4.7AHH25 pKa = 6.9PDD27 pKa = 3.49DD28 pKa = 6.21PIFACEE34 pKa = 3.7MTRR37 pKa = 11.84AYY39 pKa = 10.54GDD41 pKa = 4.44DD42 pKa = 4.01EE43 pKa = 6.11NDD45 pKa = 4.32DD46 pKa = 4.25ALSCNFEE53 pKa = 3.93GSGAFDD59 pKa = 3.03GAVFDD64 pKa = 4.45EE65 pKa = 5.32EE66 pKa = 5.16EE67 pKa = 4.47NEE69 pKa = 4.62CCDD72 pKa = 5.17DD73 pKa = 4.35DD74 pKa = 6.63HH75 pKa = 9.25DD76 pKa = 5.69DD77 pKa = 4.82EE78 pKa = 4.64IAKK81 pKa = 8.08VTQDD85 pKa = 3.63EE86 pKa = 4.43KK87 pKa = 11.04SGVYY91 pKa = 10.3GMSYY95 pKa = 10.89CEE97 pKa = 4.5DD98 pKa = 3.9DD99 pKa = 4.92DD100 pKa = 4.55MEE102 pKa = 4.39EE103 pKa = 4.04EE104 pKa = 4.3HH105 pKa = 6.65MKK107 pKa = 10.47SHH109 pKa = 6.27VSFDD113 pKa = 3.62SGHH116 pKa = 5.85EE117 pKa = 4.1FVDD120 pKa = 3.71EE121 pKa = 4.16MEE123 pKa = 4.39KK124 pKa = 10.71NRR126 pKa = 11.84LFWEE130 pKa = 4.55ACLASS135 pKa = 3.65
Molecular weight: 15.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L9T8I7|A0A0L9T8I7_PHAAN Cytokinin riboside 5'-monophosphate phosphoribohydrolase OS=Phaseolus angularis OX=3914 GN=LR48_Vigan345s000900 PE=3 SV=1
MM1 pKa = 7.24HH2 pKa = 7.21FVNFVHH8 pKa = 6.87IVAFVTFVNFVHH20 pKa = 7.18FIHH23 pKa = 6.56FVSSSTSSTSLFSSTLSNSSTSSTLSTWTTLSTSSNSFNSSTSSTSSISSTSSVHH78 pKa = 5.93PLPPVRR84 pKa = 11.84PLPPVRR90 pKa = 11.84SLRR93 pKa = 11.84HH94 pKa = 5.25FNHH97 pKa = 6.3FVSSSPSSSSSTSSTSSTSSTSSTSSTSAVRR128 pKa = 11.84PLRR131 pKa = 11.84PLHH134 pKa = 6.63PLRR137 pKa = 11.84QLRR140 pKa = 11.84PLRR143 pKa = 11.84RR144 pKa = 11.84LRR146 pKa = 11.84PLRR149 pKa = 11.84PIRR152 pKa = 11.84QLRR155 pKa = 11.84SLRR158 pKa = 11.84PLCPLGPLRR167 pKa = 11.84QLRR170 pKa = 11.84QLPPLPPLRR179 pKa = 11.84PLPLLHH185 pKa = 6.99PLLRR189 pKa = 11.84PLRR192 pKa = 11.84PLHH195 pKa = 6.02EE196 pKa = 4.59LRR198 pKa = 11.84PLRR201 pKa = 11.84PLPQLRR207 pKa = 11.84PFRR210 pKa = 11.84PLRR213 pKa = 11.84ALRR216 pKa = 11.84PLRR219 pKa = 11.84QLPPLRR225 pKa = 11.84PLRR228 pKa = 11.84QRR230 pKa = 11.84RR231 pKa = 11.84PLRR234 pKa = 11.84PLRR237 pKa = 11.84QFIHH241 pKa = 6.61FLQFVHH247 pKa = 7.19FIHH250 pKa = 6.6FVSFVHH256 pKa = 6.53FVSSSTSSTSSTSSTSSTSSVRR278 pKa = 11.84PLPLVRR284 pKa = 11.84LLNPLRR290 pKa = 11.84QFVHH294 pKa = 6.41FVNFVHH300 pKa = 7.17FVHH303 pKa = 6.5SLNSVHH309 pKa = 6.54FVHH312 pKa = 7.14FVHH315 pKa = 6.94FVSSSPSSTPSTSSTSATSSTSLVRR340 pKa = 11.84PLPPLRR346 pKa = 11.84PLPPLPPLRR355 pKa = 11.84PLRR358 pKa = 4.15
MM1 pKa = 7.24HH2 pKa = 7.21FVNFVHH8 pKa = 6.87IVAFVTFVNFVHH20 pKa = 7.18FIHH23 pKa = 6.56FVSSSTSSTSLFSSTLSNSSTSSTLSTWTTLSTSSNSFNSSTSSTSSISSTSSVHH78 pKa = 5.93PLPPVRR84 pKa = 11.84PLPPVRR90 pKa = 11.84SLRR93 pKa = 11.84HH94 pKa = 5.25FNHH97 pKa = 6.3FVSSSPSSSSSTSSTSSTSSTSSTSSTSAVRR128 pKa = 11.84PLRR131 pKa = 11.84PLHH134 pKa = 6.63PLRR137 pKa = 11.84QLRR140 pKa = 11.84PLRR143 pKa = 11.84RR144 pKa = 11.84LRR146 pKa = 11.84PLRR149 pKa = 11.84PIRR152 pKa = 11.84QLRR155 pKa = 11.84SLRR158 pKa = 11.84PLCPLGPLRR167 pKa = 11.84QLRR170 pKa = 11.84QLPPLPPLRR179 pKa = 11.84PLPLLHH185 pKa = 6.99PLLRR189 pKa = 11.84PLRR192 pKa = 11.84PLHH195 pKa = 6.02EE196 pKa = 4.59LRR198 pKa = 11.84PLRR201 pKa = 11.84PLPQLRR207 pKa = 11.84PFRR210 pKa = 11.84PLRR213 pKa = 11.84ALRR216 pKa = 11.84PLRR219 pKa = 11.84QLPPLRR225 pKa = 11.84PLRR228 pKa = 11.84QRR230 pKa = 11.84RR231 pKa = 11.84PLRR234 pKa = 11.84PLRR237 pKa = 11.84QFIHH241 pKa = 6.61FLQFVHH247 pKa = 7.19FIHH250 pKa = 6.6FVSFVHH256 pKa = 6.53FVSSSTSSTSSTSSTSSTSSVRR278 pKa = 11.84PLPLVRR284 pKa = 11.84LLNPLRR290 pKa = 11.84QFVHH294 pKa = 6.41FVNFVHH300 pKa = 7.17FVHH303 pKa = 6.5SLNSVHH309 pKa = 6.54FVHH312 pKa = 7.14FVHH315 pKa = 6.94FVSSSPSSTPSTSSTSATSSTSLVRR340 pKa = 11.84PLPPLRR346 pKa = 11.84PLPPLPPLRR355 pKa = 11.84PLRR358 pKa = 4.15
Molecular weight: 40.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12545473 |
49 |
9099 |
371.9 |
41.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.452 ± 0.018 | 1.83 ± 0.008 |
5.558 ± 0.035 | 7.204 ± 0.071 |
4.147 ± 0.012 | 6.429 ± 0.018 |
2.502 ± 0.008 | 5.069 ± 0.013 |
5.917 ± 0.015 | 9.359 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.387 ± 0.007 | 4.415 ± 0.015 |
4.919 ± 0.019 | 3.627 ± 0.011 |
5.419 ± 0.015 | 8.739 ± 0.025 |
4.933 ± 0.012 | 7.071 ± 0.039 |
1.301 ± 0.005 | 2.677 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
