
Trichechus manatus papillomavirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Rhopapillomavirus; Rhopapillomavirus 2
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
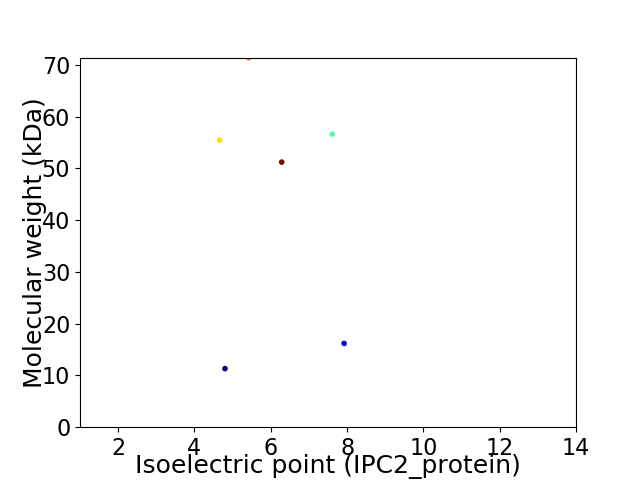
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
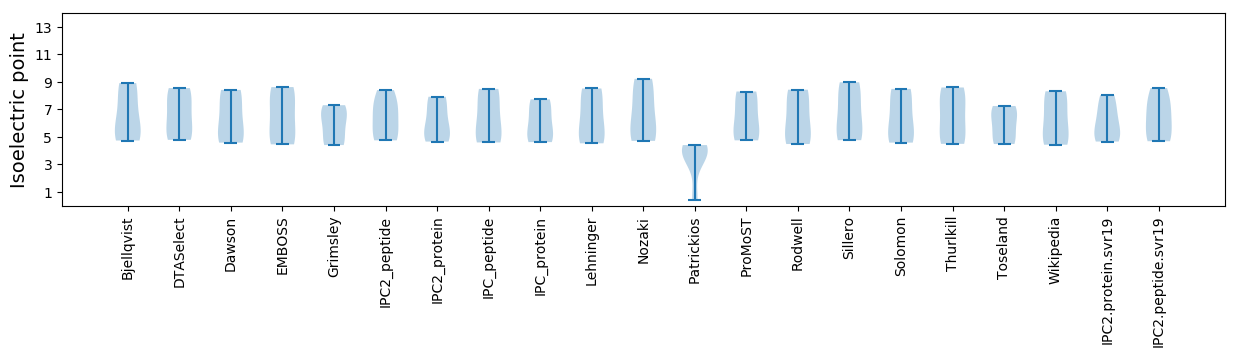
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F6TNZ5|A0A0F6TNZ5_9PAPI Replication protein E1 OS=Trichechus manatus papillomavirus 3 OX=1429799 GN=E1 PE=3 SV=1
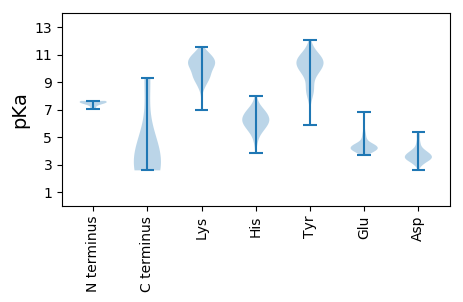
MM1 pKa = 7.49PRR3 pKa = 11.84LRR5 pKa = 11.84TKK7 pKa = 10.47RR8 pKa = 11.84ATVEE12 pKa = 4.0SIYY15 pKa = 10.46RR16 pKa = 11.84SCKK19 pKa = 8.62ATNTCSADD27 pKa = 3.44VINTVEE33 pKa = 4.05QNTLADD39 pKa = 5.17RR40 pKa = 11.84LLKK43 pKa = 9.91WISGLVYY50 pKa = 10.41FGNLGIGTAGGGGGRR65 pKa = 11.84FGYY68 pKa = 10.16GALGGTRR75 pKa = 11.84AAITPSAARR84 pKa = 11.84PPVVPEE90 pKa = 3.71TLGLLEE96 pKa = 4.28VAGPTAGEE104 pKa = 3.89IDD106 pKa = 3.73AATPSIVPLLDD117 pKa = 3.57GSTVEE122 pKa = 4.34SVNIEE127 pKa = 3.95AVAEE131 pKa = 3.99VHH133 pKa = 6.37PAPPAQGTSTLGGVAEE149 pKa = 4.52TTGSILPASDD159 pKa = 4.52PPIQTAVTHH168 pKa = 6.0TEE170 pKa = 4.17FSNPAYY176 pKa = 9.96EE177 pKa = 4.42ITTSTSDD184 pKa = 3.34SVGEE188 pKa = 4.23ISTGDD193 pKa = 3.27HH194 pKa = 5.24MVVVGGGSGQSIGEE208 pKa = 4.47EE209 pKa = 3.81IPLLQLSPDD218 pKa = 3.53NTFDD222 pKa = 3.41TSDD225 pKa = 3.44VLEE228 pKa = 4.42TEE230 pKa = 4.13FGGRR234 pKa = 11.84TSTPDD239 pKa = 3.04TQAPRR244 pKa = 11.84VRR246 pKa = 11.84GDD248 pKa = 2.87RR249 pKa = 11.84GLLYY253 pKa = 10.63SRR255 pKa = 11.84YY256 pKa = 5.88TTQQEE261 pKa = 3.9ITNPTFLSTPYY272 pKa = 11.13DD273 pKa = 3.3LVAYY277 pKa = 8.53NNPAFEE283 pKa = 4.64GTDD286 pKa = 3.31SFDD289 pKa = 3.96FPPAASPYY297 pKa = 8.38TVEE300 pKa = 4.62AAPDD304 pKa = 3.6PAFNDD309 pKa = 3.56ILHH312 pKa = 6.99LGHH315 pKa = 6.96ARR317 pKa = 11.84YY318 pKa = 8.07STTATNRR325 pKa = 11.84VRR327 pKa = 11.84VSRR330 pKa = 11.84LGTRR334 pKa = 11.84PGVRR338 pKa = 11.84TRR340 pKa = 11.84TGTLLTGRR348 pKa = 11.84THH350 pKa = 7.96FYY352 pKa = 10.99QDD354 pKa = 3.25LSSINEE360 pKa = 4.21SLEE363 pKa = 3.81LTPLSEE369 pKa = 5.58RR370 pKa = 11.84IPSSHH375 pKa = 6.15STSTSIGQSIQGYY388 pKa = 8.61PDD390 pKa = 3.87NDD392 pKa = 4.61PIIIDD397 pKa = 3.68SSFEE401 pKa = 4.32IIDD404 pKa = 4.08LNSTSAEE411 pKa = 3.95YY412 pKa = 10.95SEE414 pKa = 6.05ADD416 pKa = 3.69LLDD419 pKa = 3.88IYY421 pKa = 11.28EE422 pKa = 5.56DD423 pKa = 3.8IADD426 pKa = 4.01HH427 pKa = 6.04AQLVIGRR434 pKa = 11.84PRR436 pKa = 11.84HH437 pKa = 4.9PTTVNIPIEE446 pKa = 4.02NTIGKK451 pKa = 8.25DD452 pKa = 3.39TIIDD456 pKa = 3.65SSSGFFVEE464 pKa = 4.51VPEE467 pKa = 4.45VHH469 pKa = 6.11GTTLNPTQTDD479 pKa = 3.45RR480 pKa = 11.84DD481 pKa = 3.56GHH483 pKa = 7.04IFVYY487 pKa = 10.42SPHH490 pKa = 6.11LTSFDD495 pKa = 4.08FLPHH499 pKa = 6.59PSLLKK504 pKa = 10.22RR505 pKa = 11.84KK506 pKa = 9.5RR507 pKa = 11.84KK508 pKa = 9.65RR509 pKa = 11.84SLDD512 pKa = 3.48DD513 pKa = 4.98DD514 pKa = 4.02FTILQQ519 pKa = 3.8
MM1 pKa = 7.49PRR3 pKa = 11.84LRR5 pKa = 11.84TKK7 pKa = 10.47RR8 pKa = 11.84ATVEE12 pKa = 4.0SIYY15 pKa = 10.46RR16 pKa = 11.84SCKK19 pKa = 8.62ATNTCSADD27 pKa = 3.44VINTVEE33 pKa = 4.05QNTLADD39 pKa = 5.17RR40 pKa = 11.84LLKK43 pKa = 9.91WISGLVYY50 pKa = 10.41FGNLGIGTAGGGGGRR65 pKa = 11.84FGYY68 pKa = 10.16GALGGTRR75 pKa = 11.84AAITPSAARR84 pKa = 11.84PPVVPEE90 pKa = 3.71TLGLLEE96 pKa = 4.28VAGPTAGEE104 pKa = 3.89IDD106 pKa = 3.73AATPSIVPLLDD117 pKa = 3.57GSTVEE122 pKa = 4.34SVNIEE127 pKa = 3.95AVAEE131 pKa = 3.99VHH133 pKa = 6.37PAPPAQGTSTLGGVAEE149 pKa = 4.52TTGSILPASDD159 pKa = 4.52PPIQTAVTHH168 pKa = 6.0TEE170 pKa = 4.17FSNPAYY176 pKa = 9.96EE177 pKa = 4.42ITTSTSDD184 pKa = 3.34SVGEE188 pKa = 4.23ISTGDD193 pKa = 3.27HH194 pKa = 5.24MVVVGGGSGQSIGEE208 pKa = 4.47EE209 pKa = 3.81IPLLQLSPDD218 pKa = 3.53NTFDD222 pKa = 3.41TSDD225 pKa = 3.44VLEE228 pKa = 4.42TEE230 pKa = 4.13FGGRR234 pKa = 11.84TSTPDD239 pKa = 3.04TQAPRR244 pKa = 11.84VRR246 pKa = 11.84GDD248 pKa = 2.87RR249 pKa = 11.84GLLYY253 pKa = 10.63SRR255 pKa = 11.84YY256 pKa = 5.88TTQQEE261 pKa = 3.9ITNPTFLSTPYY272 pKa = 11.13DD273 pKa = 3.3LVAYY277 pKa = 8.53NNPAFEE283 pKa = 4.64GTDD286 pKa = 3.31SFDD289 pKa = 3.96FPPAASPYY297 pKa = 8.38TVEE300 pKa = 4.62AAPDD304 pKa = 3.6PAFNDD309 pKa = 3.56ILHH312 pKa = 6.99LGHH315 pKa = 6.96ARR317 pKa = 11.84YY318 pKa = 8.07STTATNRR325 pKa = 11.84VRR327 pKa = 11.84VSRR330 pKa = 11.84LGTRR334 pKa = 11.84PGVRR338 pKa = 11.84TRR340 pKa = 11.84TGTLLTGRR348 pKa = 11.84THH350 pKa = 7.96FYY352 pKa = 10.99QDD354 pKa = 3.25LSSINEE360 pKa = 4.21SLEE363 pKa = 3.81LTPLSEE369 pKa = 5.58RR370 pKa = 11.84IPSSHH375 pKa = 6.15STSTSIGQSIQGYY388 pKa = 8.61PDD390 pKa = 3.87NDD392 pKa = 4.61PIIIDD397 pKa = 3.68SSFEE401 pKa = 4.32IIDD404 pKa = 4.08LNSTSAEE411 pKa = 3.95YY412 pKa = 10.95SEE414 pKa = 6.05ADD416 pKa = 3.69LLDD419 pKa = 3.88IYY421 pKa = 11.28EE422 pKa = 5.56DD423 pKa = 3.8IADD426 pKa = 4.01HH427 pKa = 6.04AQLVIGRR434 pKa = 11.84PRR436 pKa = 11.84HH437 pKa = 4.9PTTVNIPIEE446 pKa = 4.02NTIGKK451 pKa = 8.25DD452 pKa = 3.39TIIDD456 pKa = 3.65SSSGFFVEE464 pKa = 4.51VPEE467 pKa = 4.45VHH469 pKa = 6.11GTTLNPTQTDD479 pKa = 3.45RR480 pKa = 11.84DD481 pKa = 3.56GHH483 pKa = 7.04IFVYY487 pKa = 10.42SPHH490 pKa = 6.11LTSFDD495 pKa = 4.08FLPHH499 pKa = 6.59PSLLKK504 pKa = 10.22RR505 pKa = 11.84KK506 pKa = 9.5RR507 pKa = 11.84KK508 pKa = 9.65RR509 pKa = 11.84SLDD512 pKa = 3.48DD513 pKa = 4.98DD514 pKa = 4.02FTILQQ519 pKa = 3.8
Molecular weight: 55.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F6RB75|A0A0F6RB75_9PAPI Regulatory protein E2 OS=Trichechus manatus papillomavirus 3 OX=1429799 GN=E2 PE=3 SV=1
MM1 pKa = 7.51SLPRR5 pKa = 11.84TLQSLRR11 pKa = 11.84DD12 pKa = 4.11DD13 pKa = 4.24FGLDD17 pKa = 3.12YY18 pKa = 8.47TTFYY22 pKa = 9.74IACVLCGGKK31 pKa = 10.26LGTLDD36 pKa = 4.43LMNFEE41 pKa = 4.8KK42 pKa = 11.1GRR44 pKa = 11.84FFLIWQYY51 pKa = 11.29GLPWACCKK59 pKa = 10.56FCIRR63 pKa = 11.84WWAAKK68 pKa = 10.3DD69 pKa = 3.42RR70 pKa = 11.84VANYY74 pKa = 9.93QYY76 pKa = 10.18SAPVATVEE84 pKa = 4.88AEE86 pKa = 4.43TKK88 pKa = 8.63QTLAEE93 pKa = 4.0LHH95 pKa = 5.97VRR97 pKa = 11.84CIGCLTEE104 pKa = 4.66LTYY107 pKa = 10.87LDD109 pKa = 3.59KK110 pKa = 11.35LRR112 pKa = 11.84IEE114 pKa = 4.26RR115 pKa = 11.84LGACLHH121 pKa = 6.37LCRR124 pKa = 11.84NKK126 pKa = 9.38WRR128 pKa = 11.84GWCSRR133 pKa = 11.84CQRR136 pKa = 11.84QWW138 pKa = 2.67
MM1 pKa = 7.51SLPRR5 pKa = 11.84TLQSLRR11 pKa = 11.84DD12 pKa = 4.11DD13 pKa = 4.24FGLDD17 pKa = 3.12YY18 pKa = 8.47TTFYY22 pKa = 9.74IACVLCGGKK31 pKa = 10.26LGTLDD36 pKa = 4.43LMNFEE41 pKa = 4.8KK42 pKa = 11.1GRR44 pKa = 11.84FFLIWQYY51 pKa = 11.29GLPWACCKK59 pKa = 10.56FCIRR63 pKa = 11.84WWAAKK68 pKa = 10.3DD69 pKa = 3.42RR70 pKa = 11.84VANYY74 pKa = 9.93QYY76 pKa = 10.18SAPVATVEE84 pKa = 4.88AEE86 pKa = 4.43TKK88 pKa = 8.63QTLAEE93 pKa = 4.0LHH95 pKa = 5.97VRR97 pKa = 11.84CIGCLTEE104 pKa = 4.66LTYY107 pKa = 10.87LDD109 pKa = 3.59KK110 pKa = 11.35LRR112 pKa = 11.84IEE114 pKa = 4.26RR115 pKa = 11.84LGACLHH121 pKa = 6.37LCRR124 pKa = 11.84NKK126 pKa = 9.38WRR128 pKa = 11.84GWCSRR133 pKa = 11.84CQRR136 pKa = 11.84QWW138 pKa = 2.67
Molecular weight: 16.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2337 |
99 |
628 |
389.5 |
43.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.033 ± 0.461 | 2.525 ± 1.092 |
6.376 ± 0.367 | 5.991 ± 0.468 |
3.894 ± 0.418 | 6.675 ± 0.796 |
2.182 ± 0.143 | 4.707 ± 0.606 |
4.108 ± 0.801 | 8.772 ± 0.643 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.369 ± 0.328 | 3.894 ± 0.553 |
6.675 ± 1.083 | 4.279 ± 0.594 |
6.59 ± 0.57 | 7.274 ± 0.818 |
7.745 ± 1.183 | 5.777 ± 0.33 |
1.669 ± 0.574 | 3.466 ± 0.359 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
