
Anelloviridae sp.
Taxonomy: Viruses; Anelloviridae; unclassified Anelloviridae
Average proteome isoelectric point is 8.65
Get precalculated fractions of proteins
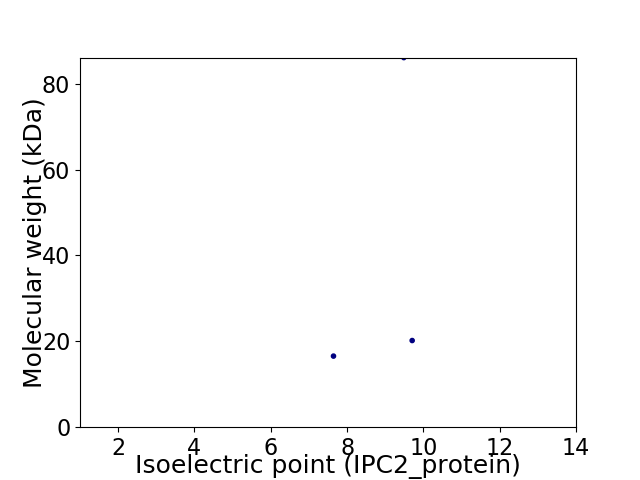
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A385E3J9|A0A385E3J9_9VIRU Capsid protein OS=Anelloviridae sp. OX=2055263 PE=3 SV=1
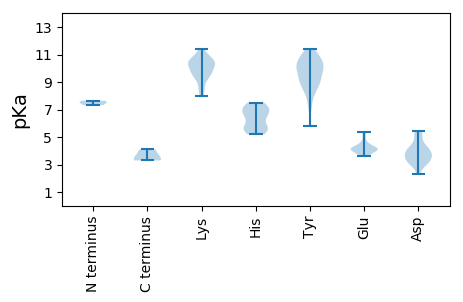
MM1 pKa = 7.51GKK3 pKa = 9.86ALKK6 pKa = 10.31KK7 pKa = 11.01AMFLGKK13 pKa = 10.33LYY15 pKa = 10.33RR16 pKa = 11.84KK17 pKa = 9.49KK18 pKa = 10.54RR19 pKa = 11.84RR20 pKa = 11.84LSLSSLHH27 pKa = 6.43CAPKK31 pKa = 10.29ACKK34 pKa = 9.59LLKK37 pKa = 10.73GMWRR41 pKa = 11.84PPTQNVSGQEE51 pKa = 4.14RR52 pKa = 11.84NWYY55 pKa = 9.37DD56 pKa = 3.14AVFYY60 pKa = 10.96SHH62 pKa = 7.26AAFCGCGDD70 pKa = 4.1CVGHH74 pKa = 7.09LSYY77 pKa = 11.07LATHH81 pKa = 7.37LGRR84 pKa = 11.84PPSAQPPPQPQPPVIRR100 pKa = 11.84RR101 pKa = 11.84LPALPAPPNPSGDD114 pKa = 3.18RR115 pKa = 11.84AAWPTGGGDD124 pKa = 3.56AGEE127 pKa = 5.05DD128 pKa = 4.04GPGEE132 pKa = 4.23GGAAGGFADD141 pKa = 5.39LADD144 pKa = 5.22DD145 pKa = 4.47EE146 pKa = 5.37LLTAAVEE153 pKa = 4.29AAEE156 pKa = 4.21EE157 pKa = 4.1
MM1 pKa = 7.51GKK3 pKa = 9.86ALKK6 pKa = 10.31KK7 pKa = 11.01AMFLGKK13 pKa = 10.33LYY15 pKa = 10.33RR16 pKa = 11.84KK17 pKa = 9.49KK18 pKa = 10.54RR19 pKa = 11.84RR20 pKa = 11.84LSLSSLHH27 pKa = 6.43CAPKK31 pKa = 10.29ACKK34 pKa = 9.59LLKK37 pKa = 10.73GMWRR41 pKa = 11.84PPTQNVSGQEE51 pKa = 4.14RR52 pKa = 11.84NWYY55 pKa = 9.37DD56 pKa = 3.14AVFYY60 pKa = 10.96SHH62 pKa = 7.26AAFCGCGDD70 pKa = 4.1CVGHH74 pKa = 7.09LSYY77 pKa = 11.07LATHH81 pKa = 7.37LGRR84 pKa = 11.84PPSAQPPPQPQPPVIRR100 pKa = 11.84RR101 pKa = 11.84LPALPAPPNPSGDD114 pKa = 3.18RR115 pKa = 11.84AAWPTGGGDD124 pKa = 3.56AGEE127 pKa = 5.05DD128 pKa = 4.04GPGEE132 pKa = 4.23GGAAGGFADD141 pKa = 5.39LADD144 pKa = 5.22DD145 pKa = 4.47EE146 pKa = 5.37LLTAAVEE153 pKa = 4.29AAEE156 pKa = 4.21EE157 pKa = 4.1
Molecular weight: 16.56 kDa
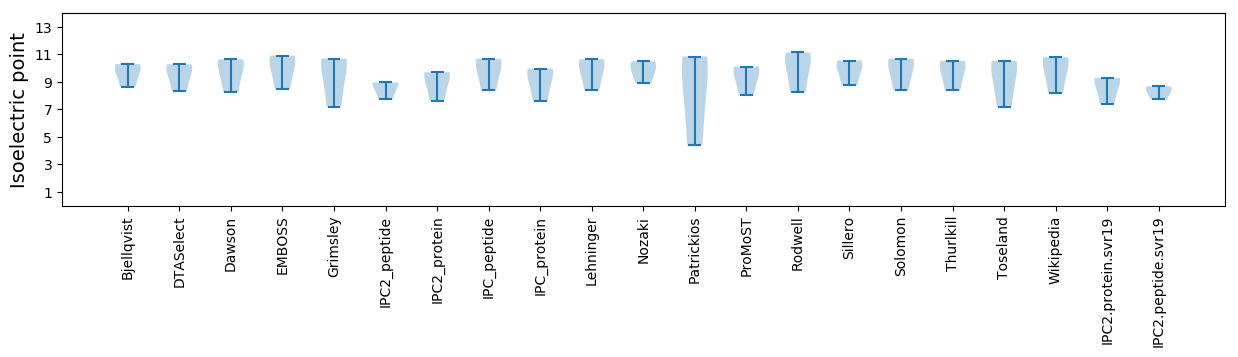
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A385E6L0|A0A385E6L0_9VIRU Uncharacterized protein OS=Anelloviridae sp. OX=2055263 PE=4 SV=1
MM1 pKa = 7.64AYY3 pKa = 8.26WWWRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PWRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84WRR21 pKa = 11.84LRR23 pKa = 11.84RR24 pKa = 11.84PRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84APYY32 pKa = 8.44RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84GRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84VRR42 pKa = 11.84KK43 pKa = 9.44RR44 pKa = 11.84RR45 pKa = 11.84GRR47 pKa = 11.84WRR49 pKa = 11.84RR50 pKa = 11.84TYY52 pKa = 10.49RR53 pKa = 11.84RR54 pKa = 11.84WRR56 pKa = 11.84RR57 pKa = 11.84KK58 pKa = 8.68RR59 pKa = 11.84GRR61 pKa = 11.84RR62 pKa = 11.84KK63 pKa = 9.46HH64 pKa = 5.55RR65 pKa = 11.84PKK67 pKa = 10.4IILKK71 pKa = 8.59QWQPNFIRR79 pKa = 11.84NCFIVGYY86 pKa = 8.81MPLIFCGEE94 pKa = 4.06NTVSQNYY101 pKa = 7.24ATHH104 pKa = 6.89SDD106 pKa = 3.67DD107 pKa = 5.47LVAKK111 pKa = 9.65GPYY114 pKa = 8.76GGGMTTTKK122 pKa = 9.14FTLRR126 pKa = 11.84ILFDD130 pKa = 3.04EE131 pKa = 4.62HH132 pKa = 7.5LRR134 pKa = 11.84YY135 pKa = 9.03MNYY138 pKa = 7.2WTVSNEE144 pKa = 3.71DD145 pKa = 3.73LDD147 pKa = 3.82LCRR150 pKa = 11.84YY151 pKa = 9.35VGAKK155 pKa = 9.93FYY157 pKa = 10.17FYY159 pKa = 10.81RR160 pKa = 11.84HH161 pKa = 5.45PTVDD165 pKa = 4.17FMVQIHH171 pKa = 5.41TQAPFLDD178 pKa = 4.23TEE180 pKa = 4.33LTGPSIHH187 pKa = 7.18PGMMILNKK195 pKa = 10.2RR196 pKa = 11.84KK197 pKa = 9.93IFVPSLKK204 pKa = 9.28TRR206 pKa = 11.84PNRR209 pKa = 11.84KK210 pKa = 8.15HH211 pKa = 5.71KK212 pKa = 10.84VRR214 pKa = 11.84VKK216 pKa = 10.46VGAPRR221 pKa = 11.84LFEE224 pKa = 5.01DD225 pKa = 2.54KK226 pKa = 10.04WYY228 pKa = 8.92PQSEE232 pKa = 4.62LCDD235 pKa = 3.6TTLVAIYY242 pKa = 8.26ATACDD247 pKa = 3.79LQFPFGSPLTEE258 pKa = 3.84NPCVNFQVLGSAYY271 pKa = 8.94KK272 pKa = 9.51QHH274 pKa = 6.97LSILTTEE281 pKa = 4.14TEE283 pKa = 4.21KK284 pKa = 11.41NKK286 pKa = 10.42AHH288 pKa = 6.5YY289 pKa = 8.99EE290 pKa = 3.68QALFNKK296 pKa = 8.35VAYY299 pKa = 10.17YY300 pKa = 9.1NTFQTIAHH308 pKa = 6.05LRR310 pKa = 11.84EE311 pKa = 3.98TGKK314 pKa = 10.02QEE316 pKa = 3.82QANPQWSTCVNSTALKK332 pKa = 10.48FDD334 pKa = 3.7GTNQNQCNDD343 pKa = 2.33TWYY346 pKa = 11.0YY347 pKa = 10.12GNAYY351 pKa = 9.47NCNIQSLCEE360 pKa = 3.97KK361 pKa = 9.77TRR363 pKa = 11.84QKK365 pKa = 10.92YY366 pKa = 9.45FKK368 pKa = 8.83ATKK371 pKa = 10.01SAFPAYY377 pKa = 8.32ATLVDD382 pKa = 5.31DD383 pKa = 4.86LYY385 pKa = 10.26EE386 pKa = 4.11YY387 pKa = 10.85HH388 pKa = 6.81GGIYY392 pKa = 10.33SSIFLSAGRR401 pKa = 11.84SYY403 pKa = 11.41FEE405 pKa = 3.91TLGAYY410 pKa = 10.15SDD412 pKa = 4.3IIYY415 pKa = 10.87NPFVDD420 pKa = 4.42KK421 pKa = 11.26GLGNIVWIDD430 pKa = 3.67YY431 pKa = 10.16LSKK434 pKa = 10.86SDD436 pKa = 5.01AVWKK440 pKa = 9.62PPQSKK445 pKa = 8.33CTIKK449 pKa = 10.72DD450 pKa = 3.37QPLWAAFNGYY460 pKa = 9.26SEE462 pKa = 5.38FCAKK466 pKa = 9.43STGDD470 pKa = 3.2GAVNINCRR478 pKa = 11.84VVVRR482 pKa = 11.84CPYY485 pKa = 5.83TTPMLIDD492 pKa = 3.91NKK494 pKa = 10.79DD495 pKa = 3.55PNQGFVPYY503 pKa = 10.41SKK505 pKa = 11.04NFGNAKK511 pKa = 9.17MKK513 pKa = 10.58GGSADD518 pKa = 3.48VPLSWRR524 pKa = 11.84VKK526 pKa = 8.01WYY528 pKa = 11.04VMMYY532 pKa = 9.13HH533 pKa = 5.21QQEE536 pKa = 4.12FMEE539 pKa = 5.36EE540 pKa = 4.1IVQSGPWAYY549 pKa = 9.95KK550 pKa = 10.45GDD552 pKa = 3.94QKK554 pKa = 11.33SAALNMKK561 pKa = 9.88YY562 pKa = 9.92VFKK565 pKa = 10.26WKK567 pKa = 9.49WGGNPISGQVVRR579 pKa = 11.84NPCKK583 pKa = 9.14TSGSSRR589 pKa = 11.84EE590 pKa = 3.89PRR592 pKa = 11.84SIQAVDD598 pKa = 4.09PKK600 pKa = 10.55RR601 pKa = 11.84VAPPLVFHH609 pKa = 6.36SWDD612 pKa = 3.15YY613 pKa = 9.77RR614 pKa = 11.84RR615 pKa = 11.84GYY617 pKa = 9.93FGHH620 pKa = 7.27ASIKK624 pKa = 10.35RR625 pKa = 11.84MSEE628 pKa = 3.68EE629 pKa = 4.34SEE631 pKa = 4.21PPTLFTGPSGKK642 pKa = 9.81RR643 pKa = 11.84PRR645 pKa = 11.84RR646 pKa = 11.84DD647 pKa = 3.15TNVQQAEE654 pKa = 4.23EE655 pKa = 4.17EE656 pKa = 4.24QQKK659 pKa = 10.34EE660 pKa = 4.11SSTFRR665 pKa = 11.84VQQQLQPWINSSQEE679 pKa = 4.11TQSSQEE685 pKa = 4.07EE686 pKa = 4.08IQAQGSIQEE695 pKa = 4.15QLLQQLGEE703 pKa = 3.9QRR705 pKa = 11.84ALRR708 pKa = 11.84KK709 pKa = 9.49QLEE712 pKa = 4.3HH713 pKa = 6.71VARR716 pKa = 11.84QVLKK720 pKa = 10.52VQAGTHH726 pKa = 5.72LHH728 pKa = 6.75PLLSSQAA735 pKa = 3.44
MM1 pKa = 7.64AYY3 pKa = 8.26WWWRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84PWRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84WRR21 pKa = 11.84LRR23 pKa = 11.84RR24 pKa = 11.84PRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84APYY32 pKa = 8.44RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84GRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84VRR42 pKa = 11.84KK43 pKa = 9.44RR44 pKa = 11.84RR45 pKa = 11.84GRR47 pKa = 11.84WRR49 pKa = 11.84RR50 pKa = 11.84TYY52 pKa = 10.49RR53 pKa = 11.84RR54 pKa = 11.84WRR56 pKa = 11.84RR57 pKa = 11.84KK58 pKa = 8.68RR59 pKa = 11.84GRR61 pKa = 11.84RR62 pKa = 11.84KK63 pKa = 9.46HH64 pKa = 5.55RR65 pKa = 11.84PKK67 pKa = 10.4IILKK71 pKa = 8.59QWQPNFIRR79 pKa = 11.84NCFIVGYY86 pKa = 8.81MPLIFCGEE94 pKa = 4.06NTVSQNYY101 pKa = 7.24ATHH104 pKa = 6.89SDD106 pKa = 3.67DD107 pKa = 5.47LVAKK111 pKa = 9.65GPYY114 pKa = 8.76GGGMTTTKK122 pKa = 9.14FTLRR126 pKa = 11.84ILFDD130 pKa = 3.04EE131 pKa = 4.62HH132 pKa = 7.5LRR134 pKa = 11.84YY135 pKa = 9.03MNYY138 pKa = 7.2WTVSNEE144 pKa = 3.71DD145 pKa = 3.73LDD147 pKa = 3.82LCRR150 pKa = 11.84YY151 pKa = 9.35VGAKK155 pKa = 9.93FYY157 pKa = 10.17FYY159 pKa = 10.81RR160 pKa = 11.84HH161 pKa = 5.45PTVDD165 pKa = 4.17FMVQIHH171 pKa = 5.41TQAPFLDD178 pKa = 4.23TEE180 pKa = 4.33LTGPSIHH187 pKa = 7.18PGMMILNKK195 pKa = 10.2RR196 pKa = 11.84KK197 pKa = 9.93IFVPSLKK204 pKa = 9.28TRR206 pKa = 11.84PNRR209 pKa = 11.84KK210 pKa = 8.15HH211 pKa = 5.71KK212 pKa = 10.84VRR214 pKa = 11.84VKK216 pKa = 10.46VGAPRR221 pKa = 11.84LFEE224 pKa = 5.01DD225 pKa = 2.54KK226 pKa = 10.04WYY228 pKa = 8.92PQSEE232 pKa = 4.62LCDD235 pKa = 3.6TTLVAIYY242 pKa = 8.26ATACDD247 pKa = 3.79LQFPFGSPLTEE258 pKa = 3.84NPCVNFQVLGSAYY271 pKa = 8.94KK272 pKa = 9.51QHH274 pKa = 6.97LSILTTEE281 pKa = 4.14TEE283 pKa = 4.21KK284 pKa = 11.41NKK286 pKa = 10.42AHH288 pKa = 6.5YY289 pKa = 8.99EE290 pKa = 3.68QALFNKK296 pKa = 8.35VAYY299 pKa = 10.17YY300 pKa = 9.1NTFQTIAHH308 pKa = 6.05LRR310 pKa = 11.84EE311 pKa = 3.98TGKK314 pKa = 10.02QEE316 pKa = 3.82QANPQWSTCVNSTALKK332 pKa = 10.48FDD334 pKa = 3.7GTNQNQCNDD343 pKa = 2.33TWYY346 pKa = 11.0YY347 pKa = 10.12GNAYY351 pKa = 9.47NCNIQSLCEE360 pKa = 3.97KK361 pKa = 9.77TRR363 pKa = 11.84QKK365 pKa = 10.92YY366 pKa = 9.45FKK368 pKa = 8.83ATKK371 pKa = 10.01SAFPAYY377 pKa = 8.32ATLVDD382 pKa = 5.31DD383 pKa = 4.86LYY385 pKa = 10.26EE386 pKa = 4.11YY387 pKa = 10.85HH388 pKa = 6.81GGIYY392 pKa = 10.33SSIFLSAGRR401 pKa = 11.84SYY403 pKa = 11.41FEE405 pKa = 3.91TLGAYY410 pKa = 10.15SDD412 pKa = 4.3IIYY415 pKa = 10.87NPFVDD420 pKa = 4.42KK421 pKa = 11.26GLGNIVWIDD430 pKa = 3.67YY431 pKa = 10.16LSKK434 pKa = 10.86SDD436 pKa = 5.01AVWKK440 pKa = 9.62PPQSKK445 pKa = 8.33CTIKK449 pKa = 10.72DD450 pKa = 3.37QPLWAAFNGYY460 pKa = 9.26SEE462 pKa = 5.38FCAKK466 pKa = 9.43STGDD470 pKa = 3.2GAVNINCRR478 pKa = 11.84VVVRR482 pKa = 11.84CPYY485 pKa = 5.83TTPMLIDD492 pKa = 3.91NKK494 pKa = 10.79DD495 pKa = 3.55PNQGFVPYY503 pKa = 10.41SKK505 pKa = 11.04NFGNAKK511 pKa = 9.17MKK513 pKa = 10.58GGSADD518 pKa = 3.48VPLSWRR524 pKa = 11.84VKK526 pKa = 8.01WYY528 pKa = 11.04VMMYY532 pKa = 9.13HH533 pKa = 5.21QQEE536 pKa = 4.12FMEE539 pKa = 5.36EE540 pKa = 4.1IVQSGPWAYY549 pKa = 9.95KK550 pKa = 10.45GDD552 pKa = 3.94QKK554 pKa = 11.33SAALNMKK561 pKa = 9.88YY562 pKa = 9.92VFKK565 pKa = 10.26WKK567 pKa = 9.49WGGNPISGQVVRR579 pKa = 11.84NPCKK583 pKa = 9.14TSGSSRR589 pKa = 11.84EE590 pKa = 3.89PRR592 pKa = 11.84SIQAVDD598 pKa = 4.09PKK600 pKa = 10.55RR601 pKa = 11.84VAPPLVFHH609 pKa = 6.36SWDD612 pKa = 3.15YY613 pKa = 9.77RR614 pKa = 11.84RR615 pKa = 11.84GYY617 pKa = 9.93FGHH620 pKa = 7.27ASIKK624 pKa = 10.35RR625 pKa = 11.84MSEE628 pKa = 3.68EE629 pKa = 4.34SEE631 pKa = 4.21PPTLFTGPSGKK642 pKa = 9.81RR643 pKa = 11.84PRR645 pKa = 11.84RR646 pKa = 11.84DD647 pKa = 3.15TNVQQAEE654 pKa = 4.23EE655 pKa = 4.17EE656 pKa = 4.24QQKK659 pKa = 10.34EE660 pKa = 4.11SSTFRR665 pKa = 11.84VQQQLQPWINSSQEE679 pKa = 4.11TQSSQEE685 pKa = 4.07EE686 pKa = 4.08IQAQGSIQEE695 pKa = 4.15QLLQQLGEE703 pKa = 3.9QRR705 pKa = 11.84ALRR708 pKa = 11.84KK709 pKa = 9.49QLEE712 pKa = 4.3HH713 pKa = 6.71VARR716 pKa = 11.84QVLKK720 pKa = 10.52VQAGTHH726 pKa = 5.72LHH728 pKa = 6.75PLLSSQAA735 pKa = 3.44
Molecular weight: 86.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1072 |
157 |
735 |
357.3 |
40.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.09 ± 1.812 | 2.052 ± 0.329 |
3.638 ± 0.465 | 4.664 ± 0.284 |
3.545 ± 1.004 | 6.437 ± 1.436 |
2.239 ± 0.174 | 2.985 ± 1.291 |
7.09 ± 1.068 | 7.183 ± 0.961 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.679 ± 0.361 | 4.291 ± 0.646 |
6.81 ± 1.383 | 5.877 ± 0.94 |
8.769 ± 0.835 | 8.116 ± 3.021 |
5.504 ± 0.757 | 4.944 ± 0.741 |
2.425 ± 0.725 | 4.664 ± 0.635 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
