
Magnaporthe grisea (Crabgrass-specific blast fungus) (Pyricularia grisea)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Sordariomycetidae;
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
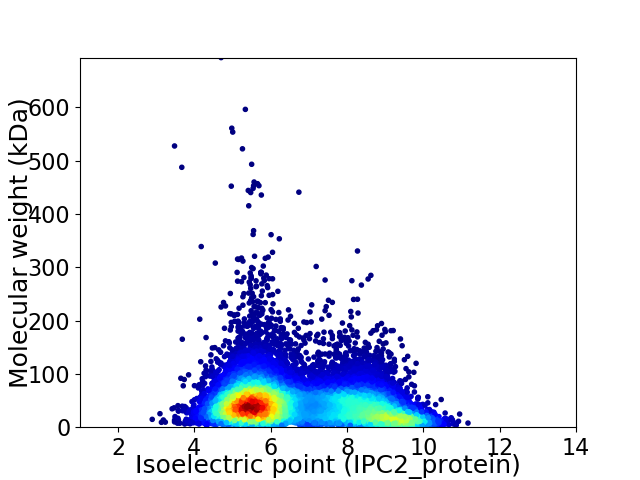
Virtual 2D-PAGE plot for 12455 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
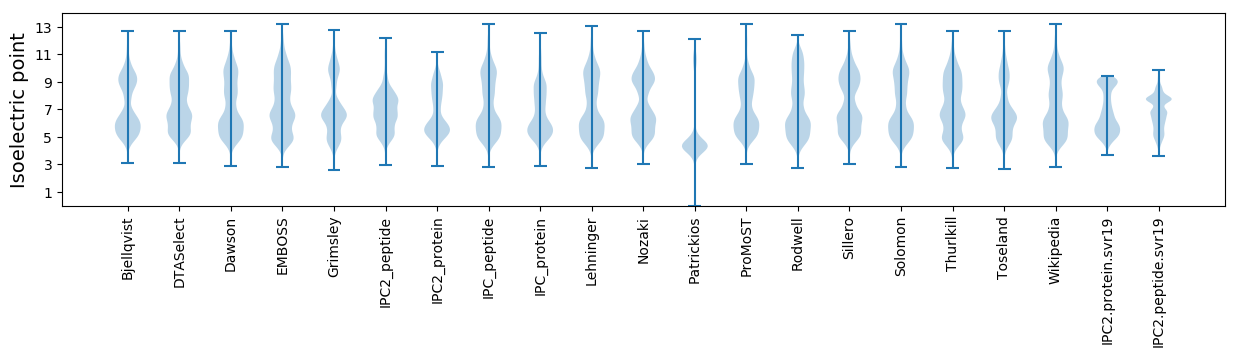
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P8B9F8|A0A6P8B9F8_MAGGR uncharacterized protein OS=Magnaporthe grisea OX=148305 GN=PgNI_02904 PE=3 SV=1
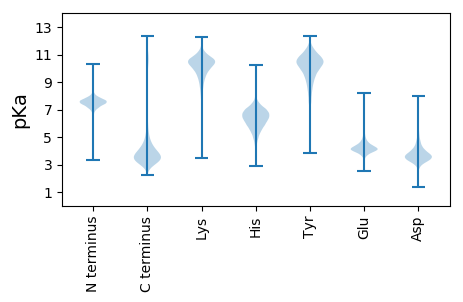
MM1 pKa = 7.58PNKK4 pKa = 10.39SQMLLVLAAALVPLAWGGSSSSILRR29 pKa = 11.84PRR31 pKa = 11.84QQGTQDD37 pKa = 3.22MPRR40 pKa = 11.84PTFGPVPWGEE50 pKa = 3.7QLGRR54 pKa = 11.84DD55 pKa = 4.1DD56 pKa = 5.18FPCARR61 pKa = 11.84AGQIAFTFEE70 pKa = 5.2DD71 pKa = 4.98GPSPYY76 pKa = 9.79TPSILNTLSQYY87 pKa = 11.39NIIASFFMTGSFEE100 pKa = 5.25DD101 pKa = 3.94GGLAQPRR108 pKa = 11.84YY109 pKa = 9.19HH110 pKa = 6.41GLARR114 pKa = 11.84FMYY117 pKa = 9.86NQGHH121 pKa = 6.88LLGSHH126 pKa = 6.63SYY128 pKa = 10.44SHH130 pKa = 7.93DD131 pKa = 3.47DD132 pKa = 3.23LRR134 pKa = 11.84SFTEE138 pKa = 3.95EE139 pKa = 3.66EE140 pKa = 3.98VRR142 pKa = 11.84ADD144 pKa = 3.94LLEE147 pKa = 4.84LEE149 pKa = 4.41ATFADD154 pKa = 3.91VLGVVPTYY162 pKa = 10.36FRR164 pKa = 11.84APFTEE169 pKa = 4.84CDD171 pKa = 3.49VGDD174 pKa = 4.08CLVNIRR180 pKa = 11.84PLVYY184 pKa = 10.41HH185 pKa = 6.51IVDD188 pKa = 3.66YY189 pKa = 11.47NIDD192 pKa = 3.62SFDD195 pKa = 3.86SYY197 pKa = 11.64EE198 pKa = 4.19LADD201 pKa = 4.23PADD204 pKa = 3.38LVAFFRR210 pKa = 11.84EE211 pKa = 4.25TMEE214 pKa = 4.36NADD217 pKa = 4.2PAVASYY223 pKa = 10.44IVRR226 pKa = 11.84FHH228 pKa = 6.71DD229 pKa = 3.83TEE231 pKa = 4.19EE232 pKa = 4.11VTANGLLEE240 pKa = 4.16EE241 pKa = 5.47FILIAATRR249 pKa = 11.84GFEE252 pKa = 3.95FVTVGEE258 pKa = 4.74CLGDD262 pKa = 3.77DD263 pKa = 3.62PGNFYY268 pKa = 10.97RR269 pKa = 11.84NPQTGNALGNGNEE282 pKa = 4.69PPPSPDD288 pKa = 3.18PSSSTEE294 pKa = 3.85STAVSASTSVSSSSVDD310 pKa = 3.51LTSSAIEE317 pKa = 4.02TTGTPEE323 pKa = 3.41ITGNPDD329 pKa = 2.92SMVSDD334 pKa = 4.11TQTSTATDD342 pKa = 3.4QEE344 pKa = 4.52VTITLDD350 pKa = 3.14TLTQEE355 pKa = 4.53TPSVASTTIDD365 pKa = 3.17TAGSDD370 pKa = 3.9DD371 pKa = 3.72PTITEE376 pKa = 4.41TQTITQTEE384 pKa = 4.5TDD386 pKa = 3.37TADD389 pKa = 3.46FPGATTTPAVAGTSDD404 pKa = 3.58MVTTSDD410 pKa = 3.47STPTITEE417 pKa = 3.67ISVSSVADD425 pKa = 3.45VTSSEE430 pKa = 4.25EE431 pKa = 4.23TTSDD435 pKa = 3.08ASQTALEE442 pKa = 4.38SSLDD446 pKa = 3.67ATSSLVTTEE455 pKa = 4.68LASQTTAAATSEE467 pKa = 4.4EE468 pKa = 4.28PSASHH473 pKa = 6.7SLITMSIISEE483 pKa = 4.37TTSTGFDD490 pKa = 3.48TPSSSVVDD498 pKa = 4.14GQTTSGAPSDD508 pKa = 4.98DD509 pKa = 4.81GITTSTSTAPTSEE522 pKa = 4.3VPTSEE527 pKa = 3.87VTDD530 pKa = 3.5TGFCTSTASFAGSLVPSDD548 pKa = 3.86TVSDD552 pKa = 3.46ISTPTATITTTCSTATCTTAATTSADD578 pKa = 3.27EE579 pKa = 4.3TAPSVTSTDD588 pKa = 3.19SGSQSSAATNFPHH601 pKa = 7.53SIITVVTKK609 pKa = 10.07TLDD612 pKa = 3.3STTQDD617 pKa = 2.9AQTNSQNTPSPSSSISPSDD636 pKa = 3.63VTPLPLSSSTEE647 pKa = 3.66IRR649 pKa = 11.84DD650 pKa = 3.3SSTIEE655 pKa = 4.11EE656 pKa = 4.48DD657 pKa = 3.46TSSATAMPNSASYY670 pKa = 7.92TTFPHH675 pKa = 5.77QFSSAEE681 pKa = 4.09VQSTGAASTTINDD694 pKa = 3.13QDD696 pKa = 3.96TNEE699 pKa = 4.2TANPATHH706 pKa = 5.42THH708 pKa = 6.12YY709 pKa = 10.95GSSTTITHH717 pKa = 6.17TAGATDD723 pKa = 4.61CPTATAANDD732 pKa = 4.02DD733 pKa = 4.26AQQSPPTPTEE743 pKa = 3.92TPQNGGGPGQEE754 pKa = 3.81TSSRR758 pKa = 11.84CGGCQDD764 pKa = 3.51GVVGGEE770 pKa = 3.94GSFGNWLTVTRR781 pKa = 11.84SSAIGRR787 pKa = 11.84DD788 pKa = 3.84DD789 pKa = 3.92LQSAGTSAGGISAGVGIQTYY809 pKa = 9.8VISSAQPPSASVAGQGAGEE828 pKa = 4.34LPSGAEE834 pKa = 3.89TGGVGGSSAVPTRR847 pKa = 11.84VVGAGGFAVGASGCAAVFAAVMAWMVLL874 pKa = 3.56
MM1 pKa = 7.58PNKK4 pKa = 10.39SQMLLVLAAALVPLAWGGSSSSILRR29 pKa = 11.84PRR31 pKa = 11.84QQGTQDD37 pKa = 3.22MPRR40 pKa = 11.84PTFGPVPWGEE50 pKa = 3.7QLGRR54 pKa = 11.84DD55 pKa = 4.1DD56 pKa = 5.18FPCARR61 pKa = 11.84AGQIAFTFEE70 pKa = 5.2DD71 pKa = 4.98GPSPYY76 pKa = 9.79TPSILNTLSQYY87 pKa = 11.39NIIASFFMTGSFEE100 pKa = 5.25DD101 pKa = 3.94GGLAQPRR108 pKa = 11.84YY109 pKa = 9.19HH110 pKa = 6.41GLARR114 pKa = 11.84FMYY117 pKa = 9.86NQGHH121 pKa = 6.88LLGSHH126 pKa = 6.63SYY128 pKa = 10.44SHH130 pKa = 7.93DD131 pKa = 3.47DD132 pKa = 3.23LRR134 pKa = 11.84SFTEE138 pKa = 3.95EE139 pKa = 3.66EE140 pKa = 3.98VRR142 pKa = 11.84ADD144 pKa = 3.94LLEE147 pKa = 4.84LEE149 pKa = 4.41ATFADD154 pKa = 3.91VLGVVPTYY162 pKa = 10.36FRR164 pKa = 11.84APFTEE169 pKa = 4.84CDD171 pKa = 3.49VGDD174 pKa = 4.08CLVNIRR180 pKa = 11.84PLVYY184 pKa = 10.41HH185 pKa = 6.51IVDD188 pKa = 3.66YY189 pKa = 11.47NIDD192 pKa = 3.62SFDD195 pKa = 3.86SYY197 pKa = 11.64EE198 pKa = 4.19LADD201 pKa = 4.23PADD204 pKa = 3.38LVAFFRR210 pKa = 11.84EE211 pKa = 4.25TMEE214 pKa = 4.36NADD217 pKa = 4.2PAVASYY223 pKa = 10.44IVRR226 pKa = 11.84FHH228 pKa = 6.71DD229 pKa = 3.83TEE231 pKa = 4.19EE232 pKa = 4.11VTANGLLEE240 pKa = 4.16EE241 pKa = 5.47FILIAATRR249 pKa = 11.84GFEE252 pKa = 3.95FVTVGEE258 pKa = 4.74CLGDD262 pKa = 3.77DD263 pKa = 3.62PGNFYY268 pKa = 10.97RR269 pKa = 11.84NPQTGNALGNGNEE282 pKa = 4.69PPPSPDD288 pKa = 3.18PSSSTEE294 pKa = 3.85STAVSASTSVSSSSVDD310 pKa = 3.51LTSSAIEE317 pKa = 4.02TTGTPEE323 pKa = 3.41ITGNPDD329 pKa = 2.92SMVSDD334 pKa = 4.11TQTSTATDD342 pKa = 3.4QEE344 pKa = 4.52VTITLDD350 pKa = 3.14TLTQEE355 pKa = 4.53TPSVASTTIDD365 pKa = 3.17TAGSDD370 pKa = 3.9DD371 pKa = 3.72PTITEE376 pKa = 4.41TQTITQTEE384 pKa = 4.5TDD386 pKa = 3.37TADD389 pKa = 3.46FPGATTTPAVAGTSDD404 pKa = 3.58MVTTSDD410 pKa = 3.47STPTITEE417 pKa = 3.67ISVSSVADD425 pKa = 3.45VTSSEE430 pKa = 4.25EE431 pKa = 4.23TTSDD435 pKa = 3.08ASQTALEE442 pKa = 4.38SSLDD446 pKa = 3.67ATSSLVTTEE455 pKa = 4.68LASQTTAAATSEE467 pKa = 4.4EE468 pKa = 4.28PSASHH473 pKa = 6.7SLITMSIISEE483 pKa = 4.37TTSTGFDD490 pKa = 3.48TPSSSVVDD498 pKa = 4.14GQTTSGAPSDD508 pKa = 4.98DD509 pKa = 4.81GITTSTSTAPTSEE522 pKa = 4.3VPTSEE527 pKa = 3.87VTDD530 pKa = 3.5TGFCTSTASFAGSLVPSDD548 pKa = 3.86TVSDD552 pKa = 3.46ISTPTATITTTCSTATCTTAATTSADD578 pKa = 3.27EE579 pKa = 4.3TAPSVTSTDD588 pKa = 3.19SGSQSSAATNFPHH601 pKa = 7.53SIITVVTKK609 pKa = 10.07TLDD612 pKa = 3.3STTQDD617 pKa = 2.9AQTNSQNTPSPSSSISPSDD636 pKa = 3.63VTPLPLSSSTEE647 pKa = 3.66IRR649 pKa = 11.84DD650 pKa = 3.3SSTIEE655 pKa = 4.11EE656 pKa = 4.48DD657 pKa = 3.46TSSATAMPNSASYY670 pKa = 7.92TTFPHH675 pKa = 5.77QFSSAEE681 pKa = 4.09VQSTGAASTTINDD694 pKa = 3.13QDD696 pKa = 3.96TNEE699 pKa = 4.2TANPATHH706 pKa = 5.42THH708 pKa = 6.12YY709 pKa = 10.95GSSTTITHH717 pKa = 6.17TAGATDD723 pKa = 4.61CPTATAANDD732 pKa = 4.02DD733 pKa = 4.26AQQSPPTPTEE743 pKa = 3.92TPQNGGGPGQEE754 pKa = 3.81TSSRR758 pKa = 11.84CGGCQDD764 pKa = 3.51GVVGGEE770 pKa = 3.94GSFGNWLTVTRR781 pKa = 11.84SSAIGRR787 pKa = 11.84DD788 pKa = 3.84DD789 pKa = 3.92LQSAGTSAGGISAGVGIQTYY809 pKa = 9.8VISSAQPPSASVAGQGAGEE828 pKa = 4.34LPSGAEE834 pKa = 3.89TGGVGGSSAVPTRR847 pKa = 11.84VVGAGGFAVGASGCAAVFAAVMAWMVLL874 pKa = 3.56
Molecular weight: 89.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P8BIR3|A0A6P8BIR3_MAGGR Mannosyltransferase OS=Magnaporthe grisea OX=148305 GN=PgNI_01330 PE=3 SV=1
MM1 pKa = 7.26VFQSRR6 pKa = 11.84LRR8 pKa = 11.84FFPRR12 pKa = 11.84PGSHH16 pKa = 6.49KK17 pKa = 10.62SFRR20 pKa = 11.84TKK22 pKa = 10.45QKK24 pKa = 9.84LAKK27 pKa = 9.55AQKK30 pKa = 8.59QNRR33 pKa = 11.84PIPQWIRR40 pKa = 11.84LRR42 pKa = 11.84TGNTIRR48 pKa = 11.84YY49 pKa = 5.79NAKK52 pKa = 8.89RR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 4.14WRR57 pKa = 11.84KK58 pKa = 7.41TRR60 pKa = 11.84LGII63 pKa = 4.46
MM1 pKa = 7.26VFQSRR6 pKa = 11.84LRR8 pKa = 11.84FFPRR12 pKa = 11.84PGSHH16 pKa = 6.49KK17 pKa = 10.62SFRR20 pKa = 11.84TKK22 pKa = 10.45QKK24 pKa = 9.84LAKK27 pKa = 9.55AQKK30 pKa = 8.59QNRR33 pKa = 11.84PIPQWIRR40 pKa = 11.84LRR42 pKa = 11.84TGNTIRR48 pKa = 11.84YY49 pKa = 5.79NAKK52 pKa = 8.89RR53 pKa = 11.84RR54 pKa = 11.84HH55 pKa = 4.14WRR57 pKa = 11.84KK58 pKa = 7.41TRR60 pKa = 11.84LGII63 pKa = 4.46
Molecular weight: 7.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5706119 |
31 |
6480 |
458.1 |
50.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.221 ± 0.027 | 1.252 ± 0.01 |
5.761 ± 0.019 | 5.886 ± 0.025 |
3.513 ± 0.013 | 7.322 ± 0.024 |
2.325 ± 0.011 | 4.502 ± 0.015 |
4.735 ± 0.022 | 8.595 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.256 ± 0.008 | 3.6 ± 0.012 |
6.225 ± 0.026 | 4.059 ± 0.02 |
6.288 ± 0.019 | 8.225 ± 0.031 |
5.984 ± 0.022 | 6.289 ± 0.018 |
1.412 ± 0.009 | 2.547 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
