
Kazachstania saulgeensis
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Saccharomycetaceae; Kazachstania
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
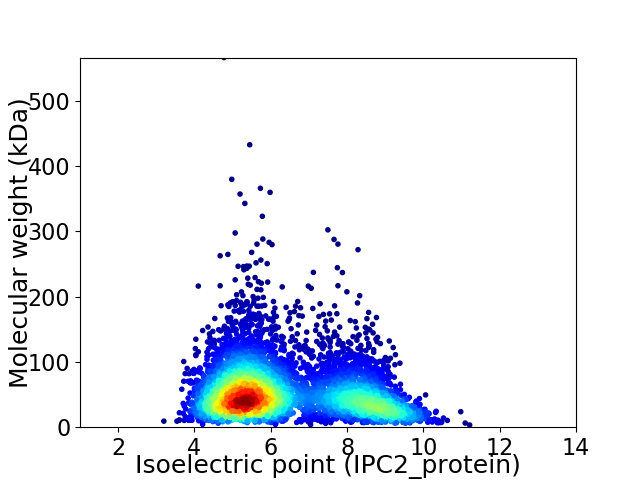
Virtual 2D-PAGE plot for 5290 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
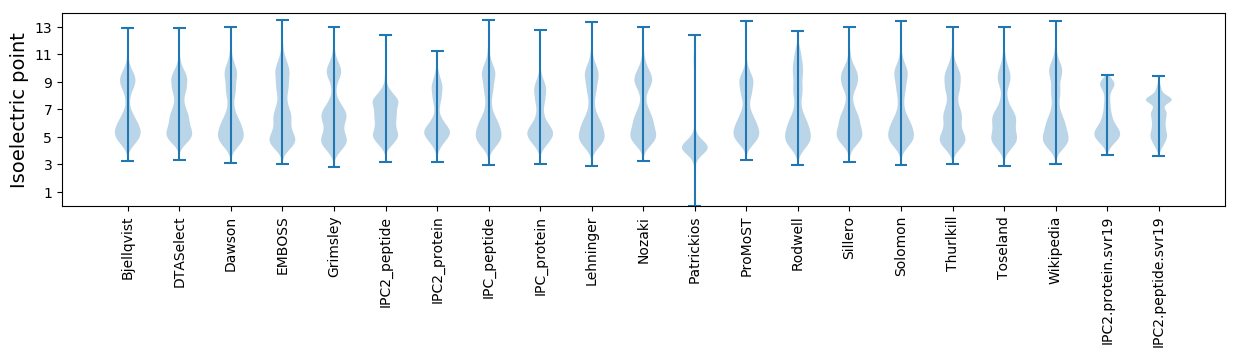
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X7R5X9|A0A1X7R5X9_9SACH Mediator complex subunit 12 OS=Kazachstania saulgeensis OX=1789683 GN=KASA_0M00275G PE=3 SV=1
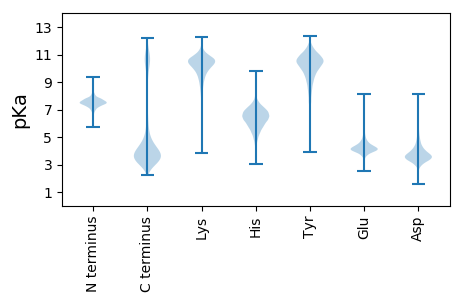
MM1 pKa = 7.45KK2 pKa = 10.5LSTVVVLLGSSSLALAALSAEE23 pKa = 4.48DD24 pKa = 3.9LAEE27 pKa = 3.99LTVIFSDD34 pKa = 3.38INKK37 pKa = 10.14NGDD40 pKa = 3.47SYY42 pKa = 11.22TNWILDD48 pKa = 3.57SGIDD52 pKa = 3.49MPMDD56 pKa = 3.6VLPILMGVEE65 pKa = 4.13TATDD69 pKa = 3.42SSYY72 pKa = 8.1TTYY75 pKa = 11.35LDD77 pKa = 3.46QLNDD81 pKa = 3.56ADD83 pKa = 3.83VSAFEE88 pKa = 4.61GVATSVPWYY97 pKa = 9.09TSYY100 pKa = 11.23LSSEE104 pKa = 4.06IAVAVANANAEE115 pKa = 4.28AEE117 pKa = 4.52TTAATSAATSSAVDD131 pKa = 3.53SSSATSSVVEE141 pKa = 4.25SSAASAAASSNAATSSAVEE160 pKa = 4.11SSAAASSNAATSSVVEE176 pKa = 4.26SSAAASSNAATAEE189 pKa = 4.13SSAATSSVASSAVAEE204 pKa = 4.38SSAASAAASSNAATSASAATDD225 pKa = 3.19ASTASSAAATDD236 pKa = 3.95ASAATSATDD245 pKa = 3.42ATSVAQASSSAASNQVSSVASSVIASSAASSNITASQEE283 pKa = 4.33TVTNTLTDD291 pKa = 3.37ISSEE295 pKa = 4.17TTTFCPEE302 pKa = 4.18SGDD305 pKa = 3.78VTATAYY311 pKa = 10.56ANSTKK316 pKa = 10.2TVTVCDD322 pKa = 4.3DD323 pKa = 3.21ACKK326 pKa = 10.28SRR328 pKa = 11.84KK329 pKa = 9.2AAEE332 pKa = 3.89ATTTINSLTTATVTKK347 pKa = 10.44CDD349 pKa = 4.03EE350 pKa = 4.23ACHH353 pKa = 5.96ASKK356 pKa = 10.67SAASAASAAAATVTNTQITTATITSCDD383 pKa = 3.87GGCHH387 pKa = 6.11ASKK390 pKa = 10.72SSASAASVASAASAASAASVAKK412 pKa = 9.94EE413 pKa = 3.73NGVAGSASTTVQVQVQTSTTSTQSIAVQNANNAIKK448 pKa = 10.44QSAGLAGLLGAVAIFLLL465 pKa = 4.37
MM1 pKa = 7.45KK2 pKa = 10.5LSTVVVLLGSSSLALAALSAEE23 pKa = 4.48DD24 pKa = 3.9LAEE27 pKa = 3.99LTVIFSDD34 pKa = 3.38INKK37 pKa = 10.14NGDD40 pKa = 3.47SYY42 pKa = 11.22TNWILDD48 pKa = 3.57SGIDD52 pKa = 3.49MPMDD56 pKa = 3.6VLPILMGVEE65 pKa = 4.13TATDD69 pKa = 3.42SSYY72 pKa = 8.1TTYY75 pKa = 11.35LDD77 pKa = 3.46QLNDD81 pKa = 3.56ADD83 pKa = 3.83VSAFEE88 pKa = 4.61GVATSVPWYY97 pKa = 9.09TSYY100 pKa = 11.23LSSEE104 pKa = 4.06IAVAVANANAEE115 pKa = 4.28AEE117 pKa = 4.52TTAATSAATSSAVDD131 pKa = 3.53SSSATSSVVEE141 pKa = 4.25SSAASAAASSNAATSSAVEE160 pKa = 4.11SSAAASSNAATSSVVEE176 pKa = 4.26SSAAASSNAATAEE189 pKa = 4.13SSAATSSVASSAVAEE204 pKa = 4.38SSAASAAASSNAATSASAATDD225 pKa = 3.19ASTASSAAATDD236 pKa = 3.95ASAATSATDD245 pKa = 3.42ATSVAQASSSAASNQVSSVASSVIASSAASSNITASQEE283 pKa = 4.33TVTNTLTDD291 pKa = 3.37ISSEE295 pKa = 4.17TTTFCPEE302 pKa = 4.18SGDD305 pKa = 3.78VTATAYY311 pKa = 10.56ANSTKK316 pKa = 10.2TVTVCDD322 pKa = 4.3DD323 pKa = 3.21ACKK326 pKa = 10.28SRR328 pKa = 11.84KK329 pKa = 9.2AAEE332 pKa = 3.89ATTTINSLTTATVTKK347 pKa = 10.44CDD349 pKa = 4.03EE350 pKa = 4.23ACHH353 pKa = 5.96ASKK356 pKa = 10.67SAASAASAAAATVTNTQITTATITSCDD383 pKa = 3.87GGCHH387 pKa = 6.11ASKK390 pKa = 10.72SSASAASVASAASAASAASVAKK412 pKa = 9.94EE413 pKa = 3.73NGVAGSASTTVQVQVQTSTTSTQSIAVQNANNAIKK448 pKa = 10.44QSAGLAGLLGAVAIFLLL465 pKa = 4.37
Molecular weight: 44.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X7QYF1|A0A1X7QYF1_9SACH Cytoplasmic tRNA 2-thiolation protein 2 OS=Kazachstania saulgeensis OX=1789683 GN=NCS2 PE=3 SV=1
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.11WRR6 pKa = 11.84KK7 pKa = 9.07KK8 pKa = 6.93RR9 pKa = 11.84TRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.48RR15 pKa = 11.84KK16 pKa = 8.41RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.19VRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 10.98
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.11WRR6 pKa = 11.84KK7 pKa = 9.07KK8 pKa = 6.93RR9 pKa = 11.84TRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.48RR15 pKa = 11.84KK16 pKa = 8.41RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.19VRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 10.98
Molecular weight: 3.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2691636 |
25 |
4952 |
508.8 |
57.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.913 ± 0.034 | 1.134 ± 0.012 |
6.172 ± 0.023 | 6.499 ± 0.034 |
4.268 ± 0.023 | 4.817 ± 0.032 |
2.027 ± 0.013 | 7.215 ± 0.031 |
7.29 ± 0.029 | 9.242 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.189 ± 0.011 | 7.067 ± 0.039 |
4.103 ± 0.022 | 3.996 ± 0.029 |
4.117 ± 0.02 | 8.794 ± 0.041 |
6.355 ± 0.033 | 5.418 ± 0.023 |
0.967 ± 0.01 | 3.419 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
