
Torque teno midi virus 13
Taxonomy: Viruses; Anelloviridae; Gammatorquevirus
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
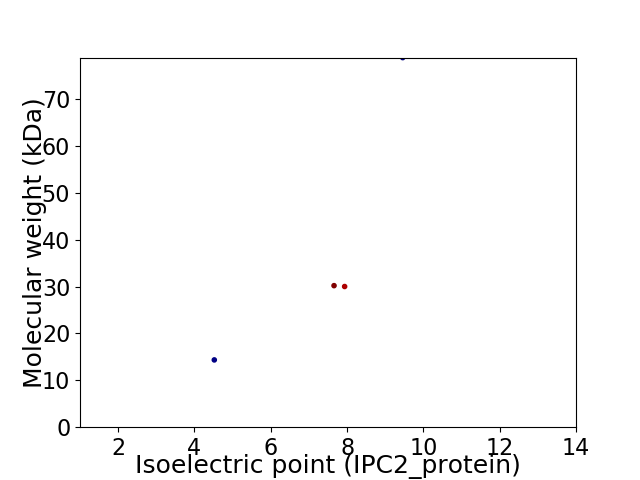
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A7VM04|A7VM04_9VIRU Capsid protein OS=Torque teno midi virus 13 OX=2065054 PE=3 SV=1
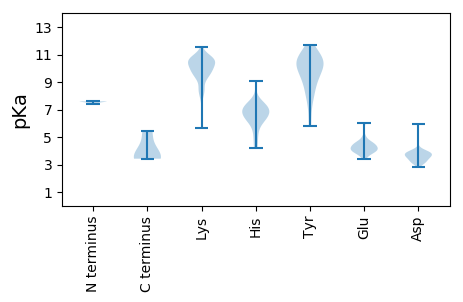
MM1 pKa = 7.6SKK3 pKa = 10.54LCSTDD8 pKa = 3.79FYY10 pKa = 11.33KK11 pKa = 9.81KK12 pKa = 9.17TPFNPEE18 pKa = 3.5TQTQIWMSQLADD30 pKa = 3.31GHH32 pKa = 7.41DD33 pKa = 3.98NFCHH37 pKa = 6.3CNSPFAHH44 pKa = 7.0LLASIFPPGHH54 pKa = 7.17KK55 pKa = 10.55DD56 pKa = 3.08RR57 pKa = 11.84DD58 pKa = 3.61LTINQILKK66 pKa = 9.78RR67 pKa = 11.84DD68 pKa = 3.9YY69 pKa = 10.88IEE71 pKa = 3.91QCRR74 pKa = 11.84STGEE78 pKa = 3.85EE79 pKa = 4.56GEE81 pKa = 4.21NSGGEE86 pKa = 4.15KK87 pKa = 9.54PDD89 pKa = 3.15QGGGFKK95 pKa = 10.74GIEE98 pKa = 3.88EE99 pKa = 4.16KK100 pKa = 10.55QQEE103 pKa = 4.51EE104 pKa = 4.47EE105 pKa = 4.71EE106 pKa = 4.15EE107 pKa = 4.03DD108 pKa = 3.84LPEE111 pKa = 6.05KK112 pKa = 10.67EE113 pKa = 4.99LIDD116 pKa = 3.78MLAAAAEE123 pKa = 4.36DD124 pKa = 3.85DD125 pKa = 3.91TRR127 pKa = 5.45
MM1 pKa = 7.6SKK3 pKa = 10.54LCSTDD8 pKa = 3.79FYY10 pKa = 11.33KK11 pKa = 9.81KK12 pKa = 9.17TPFNPEE18 pKa = 3.5TQTQIWMSQLADD30 pKa = 3.31GHH32 pKa = 7.41DD33 pKa = 3.98NFCHH37 pKa = 6.3CNSPFAHH44 pKa = 7.0LLASIFPPGHH54 pKa = 7.17KK55 pKa = 10.55DD56 pKa = 3.08RR57 pKa = 11.84DD58 pKa = 3.61LTINQILKK66 pKa = 9.78RR67 pKa = 11.84DD68 pKa = 3.9YY69 pKa = 10.88IEE71 pKa = 3.91QCRR74 pKa = 11.84STGEE78 pKa = 3.85EE79 pKa = 4.56GEE81 pKa = 4.21NSGGEE86 pKa = 4.15KK87 pKa = 9.54PDD89 pKa = 3.15QGGGFKK95 pKa = 10.74GIEE98 pKa = 3.88EE99 pKa = 4.16KK100 pKa = 10.55QQEE103 pKa = 4.51EE104 pKa = 4.47EE105 pKa = 4.71EE106 pKa = 4.15EE107 pKa = 4.03DD108 pKa = 3.84LPEE111 pKa = 6.05KK112 pKa = 10.67EE113 pKa = 4.99LIDD116 pKa = 3.78MLAAAAEE123 pKa = 4.36DD124 pKa = 3.85DD125 pKa = 3.91TRR127 pKa = 5.45
Molecular weight: 14.35 kDa
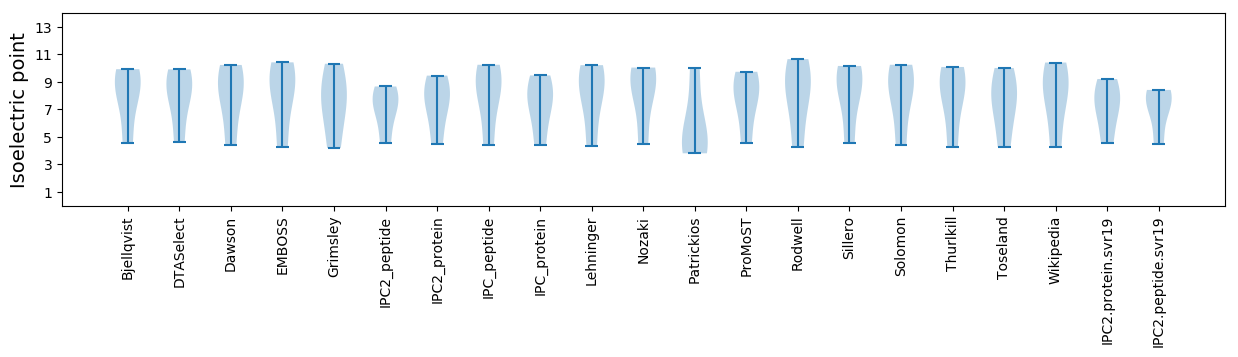
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7VM04|A7VM04_9VIRU Capsid protein OS=Torque teno midi virus 13 OX=2065054 PE=3 SV=1
MM1 pKa = 7.43PFYY4 pKa = 10.16WRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.19FWWGKK15 pKa = 9.63ARR17 pKa = 11.84PRR19 pKa = 11.84WRR21 pKa = 11.84FQRR24 pKa = 11.84YY25 pKa = 7.03RR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 5.66TTRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84PARR39 pKa = 11.84KK40 pKa = 8.65RR41 pKa = 11.84AHH43 pKa = 6.84RR44 pKa = 11.84YY45 pKa = 7.37AGRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84YY54 pKa = 8.31KK55 pKa = 10.32VRR57 pKa = 11.84RR58 pKa = 11.84KK59 pKa = 8.94RR60 pKa = 11.84QKK62 pKa = 9.76IPLFQWQPDD71 pKa = 3.98KK72 pKa = 10.98IVKK75 pKa = 10.19CKK77 pKa = 10.4IKK79 pKa = 10.73GVTTLVLGAEE89 pKa = 5.12GKK91 pKa = 9.98QLVCYY96 pKa = 10.01TNVKK100 pKa = 8.05TANTPAKK107 pKa = 10.07APGGGGFGCEE117 pKa = 3.92QFSLDD122 pKa = 4.98SLYY125 pKa = 11.21ADD127 pKa = 3.12YY128 pKa = 11.29KK129 pKa = 10.08LRR131 pKa = 11.84KK132 pKa = 9.29NVWTKK137 pKa = 11.52SNILLDD143 pKa = 3.66LCRR146 pKa = 11.84YY147 pKa = 9.39LFVKK151 pKa = 10.49ISFYY155 pKa = 10.84RR156 pKa = 11.84HH157 pKa = 6.65PEE159 pKa = 3.39TDD161 pKa = 5.92FIVTWHH167 pKa = 6.5RR168 pKa = 11.84QPPFDD173 pKa = 3.68ISKK176 pKa = 10.11EE177 pKa = 3.83IYY179 pKa = 9.63AACHH183 pKa = 5.22PQNMLLAKK191 pKa = 10.0HH192 pKa = 6.09KK193 pKa = 10.14MLILSKK199 pKa = 9.49FTNPKK204 pKa = 10.22GKK206 pKa = 9.96LKK208 pKa = 9.37KK209 pKa = 8.17TRR211 pKa = 11.84KK212 pKa = 9.27VKK214 pKa = 10.21PPKK217 pKa = 10.25QMLNKK222 pKa = 9.47WFFQEE227 pKa = 4.45HH228 pKa = 6.44FSHH231 pKa = 6.83EE232 pKa = 4.38PLLMIQATAANLNYY246 pKa = 10.23VNLGCCNTNQMCTFFYY262 pKa = 10.34INPGFYY268 pKa = 10.42QIANWGATTTTGYY281 pKa = 9.98IPYY284 pKa = 9.59SGYY287 pKa = 8.23PTATIYY293 pKa = 8.4TWNYY297 pKa = 9.04KK298 pKa = 7.54QWQTGQGGQPFTKK311 pKa = 9.66PQTYY315 pKa = 9.16NQSVNYY321 pKa = 7.97NTGFFSKK328 pKa = 10.59QMLEE332 pKa = 4.05AVQLTTDD339 pKa = 3.33QTGQRR344 pKa = 11.84PLGMNPVNICRR355 pKa = 11.84YY356 pKa = 10.26NPTLDD361 pKa = 3.51SGKK364 pKa = 10.95GNAIWLVSSHH374 pKa = 6.42ANTYY378 pKa = 10.34RR379 pKa = 11.84EE380 pKa = 4.48PITDD384 pKa = 3.12KK385 pKa = 10.93TLIFEE390 pKa = 4.51GLPLYY395 pKa = 9.72MLLYY399 pKa = 10.54GYY401 pKa = 10.6LSYY404 pKa = 10.62VQKK407 pKa = 10.78VKK409 pKa = 10.77NVRR412 pKa = 11.84DD413 pKa = 3.59FLNNYY418 pKa = 5.78TLCMRR423 pKa = 11.84SPALFPYY430 pKa = 8.92SQPGAEE436 pKa = 4.17TQVIIPVNEE445 pKa = 3.76NFIHH449 pKa = 6.92GKK451 pKa = 9.35AAYY454 pKa = 9.91DD455 pKa = 3.52EE456 pKa = 4.52TLTEE460 pKa = 4.29SVKK463 pKa = 10.94GYY465 pKa = 6.22WTPNVYY471 pKa = 9.66QQIQTLNTMVEE482 pKa = 4.06SSPYY486 pKa = 8.36VPKK489 pKa = 10.93YY490 pKa = 10.59SQTKK494 pKa = 8.75NSTWEE499 pKa = 3.46LDD501 pKa = 3.34MFYY504 pKa = 10.27TFHH507 pKa = 7.24FKK509 pKa = 10.02WGWTRR514 pKa = 11.84TNRR517 pKa = 11.84PPITDD522 pKa = 3.65PALQGTYY529 pKa = 9.19EE530 pKa = 4.34VPDD533 pKa = 3.85SLQQAVQIRR542 pKa = 11.84DD543 pKa = 3.44PGKK546 pKa = 10.23IKK548 pKa = 9.96ATSILHH554 pKa = 5.94PWDD557 pKa = 2.84IRR559 pKa = 11.84KK560 pKa = 10.05GYY562 pKa = 7.78FTTPALKK569 pKa = 10.5RR570 pKa = 11.84MSDD573 pKa = 3.38NLSIDD578 pKa = 3.4TTFQPDD584 pKa = 3.5SEE586 pKa = 4.75IIPTKK591 pKa = 10.04RR592 pKa = 11.84RR593 pKa = 11.84KK594 pKa = 8.24TGPEE598 pKa = 3.92LSTPWEE604 pKa = 4.14DD605 pKa = 3.18QKK607 pKa = 11.46EE608 pKa = 4.27IQTCLQEE615 pKa = 4.41LFKK618 pKa = 11.09KK619 pKa = 9.56STFQEE624 pKa = 4.12EE625 pKa = 4.67TEE627 pKa = 4.1EE628 pKa = 4.41DD629 pKa = 3.6LHH631 pKa = 9.07KK632 pKa = 10.84LIQQQHH638 pKa = 4.34QQQQEE643 pKa = 4.38LKK645 pKa = 8.37WNILRR650 pKa = 11.84VISEE654 pKa = 4.35IKK656 pKa = 10.23QSQNLLKK663 pKa = 10.72LQTGFPNN670 pKa = 3.45
MM1 pKa = 7.43PFYY4 pKa = 10.16WRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.19FWWGKK15 pKa = 9.63ARR17 pKa = 11.84PRR19 pKa = 11.84WRR21 pKa = 11.84FQRR24 pKa = 11.84YY25 pKa = 7.03RR26 pKa = 11.84RR27 pKa = 11.84KK28 pKa = 5.66TTRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84PARR39 pKa = 11.84KK40 pKa = 8.65RR41 pKa = 11.84AHH43 pKa = 6.84RR44 pKa = 11.84YY45 pKa = 7.37AGRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84YY54 pKa = 8.31KK55 pKa = 10.32VRR57 pKa = 11.84RR58 pKa = 11.84KK59 pKa = 8.94RR60 pKa = 11.84QKK62 pKa = 9.76IPLFQWQPDD71 pKa = 3.98KK72 pKa = 10.98IVKK75 pKa = 10.19CKK77 pKa = 10.4IKK79 pKa = 10.73GVTTLVLGAEE89 pKa = 5.12GKK91 pKa = 9.98QLVCYY96 pKa = 10.01TNVKK100 pKa = 8.05TANTPAKK107 pKa = 10.07APGGGGFGCEE117 pKa = 3.92QFSLDD122 pKa = 4.98SLYY125 pKa = 11.21ADD127 pKa = 3.12YY128 pKa = 11.29KK129 pKa = 10.08LRR131 pKa = 11.84KK132 pKa = 9.29NVWTKK137 pKa = 11.52SNILLDD143 pKa = 3.66LCRR146 pKa = 11.84YY147 pKa = 9.39LFVKK151 pKa = 10.49ISFYY155 pKa = 10.84RR156 pKa = 11.84HH157 pKa = 6.65PEE159 pKa = 3.39TDD161 pKa = 5.92FIVTWHH167 pKa = 6.5RR168 pKa = 11.84QPPFDD173 pKa = 3.68ISKK176 pKa = 10.11EE177 pKa = 3.83IYY179 pKa = 9.63AACHH183 pKa = 5.22PQNMLLAKK191 pKa = 10.0HH192 pKa = 6.09KK193 pKa = 10.14MLILSKK199 pKa = 9.49FTNPKK204 pKa = 10.22GKK206 pKa = 9.96LKK208 pKa = 9.37KK209 pKa = 8.17TRR211 pKa = 11.84KK212 pKa = 9.27VKK214 pKa = 10.21PPKK217 pKa = 10.25QMLNKK222 pKa = 9.47WFFQEE227 pKa = 4.45HH228 pKa = 6.44FSHH231 pKa = 6.83EE232 pKa = 4.38PLLMIQATAANLNYY246 pKa = 10.23VNLGCCNTNQMCTFFYY262 pKa = 10.34INPGFYY268 pKa = 10.42QIANWGATTTTGYY281 pKa = 9.98IPYY284 pKa = 9.59SGYY287 pKa = 8.23PTATIYY293 pKa = 8.4TWNYY297 pKa = 9.04KK298 pKa = 7.54QWQTGQGGQPFTKK311 pKa = 9.66PQTYY315 pKa = 9.16NQSVNYY321 pKa = 7.97NTGFFSKK328 pKa = 10.59QMLEE332 pKa = 4.05AVQLTTDD339 pKa = 3.33QTGQRR344 pKa = 11.84PLGMNPVNICRR355 pKa = 11.84YY356 pKa = 10.26NPTLDD361 pKa = 3.51SGKK364 pKa = 10.95GNAIWLVSSHH374 pKa = 6.42ANTYY378 pKa = 10.34RR379 pKa = 11.84EE380 pKa = 4.48PITDD384 pKa = 3.12KK385 pKa = 10.93TLIFEE390 pKa = 4.51GLPLYY395 pKa = 9.72MLLYY399 pKa = 10.54GYY401 pKa = 10.6LSYY404 pKa = 10.62VQKK407 pKa = 10.78VKK409 pKa = 10.77NVRR412 pKa = 11.84DD413 pKa = 3.59FLNNYY418 pKa = 5.78TLCMRR423 pKa = 11.84SPALFPYY430 pKa = 8.92SQPGAEE436 pKa = 4.17TQVIIPVNEE445 pKa = 3.76NFIHH449 pKa = 6.92GKK451 pKa = 9.35AAYY454 pKa = 9.91DD455 pKa = 3.52EE456 pKa = 4.52TLTEE460 pKa = 4.29SVKK463 pKa = 10.94GYY465 pKa = 6.22WTPNVYY471 pKa = 9.66QQIQTLNTMVEE482 pKa = 4.06SSPYY486 pKa = 8.36VPKK489 pKa = 10.93YY490 pKa = 10.59SQTKK494 pKa = 8.75NSTWEE499 pKa = 3.46LDD501 pKa = 3.34MFYY504 pKa = 10.27TFHH507 pKa = 7.24FKK509 pKa = 10.02WGWTRR514 pKa = 11.84TNRR517 pKa = 11.84PPITDD522 pKa = 3.65PALQGTYY529 pKa = 9.19EE530 pKa = 4.34VPDD533 pKa = 3.85SLQQAVQIRR542 pKa = 11.84DD543 pKa = 3.44PGKK546 pKa = 10.23IKK548 pKa = 9.96ATSILHH554 pKa = 5.94PWDD557 pKa = 2.84IRR559 pKa = 11.84KK560 pKa = 10.05GYY562 pKa = 7.78FTTPALKK569 pKa = 10.5RR570 pKa = 11.84MSDD573 pKa = 3.38NLSIDD578 pKa = 3.4TTFQPDD584 pKa = 3.5SEE586 pKa = 4.75IIPTKK591 pKa = 10.04RR592 pKa = 11.84RR593 pKa = 11.84KK594 pKa = 8.24TGPEE598 pKa = 3.92LSTPWEE604 pKa = 4.14DD605 pKa = 3.18QKK607 pKa = 11.46EE608 pKa = 4.27IQTCLQEE615 pKa = 4.41LFKK618 pKa = 11.09KK619 pKa = 9.56STFQEE624 pKa = 4.12EE625 pKa = 4.67TEE627 pKa = 4.1EE628 pKa = 4.41DD629 pKa = 3.6LHH631 pKa = 9.07KK632 pKa = 10.84LIQQQHH638 pKa = 4.34QQQQEE643 pKa = 4.38LKK645 pKa = 8.37WNILRR650 pKa = 11.84VISEE654 pKa = 4.35IKK656 pKa = 10.23QSQNLLKK663 pKa = 10.72LQTGFPNN670 pKa = 3.45
Molecular weight: 78.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1330 |
127 |
670 |
332.5 |
38.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.887 ± 0.613 | 1.805 ± 0.163 |
4.586 ± 0.86 | 6.541 ± 1.495 |
4.586 ± 0.54 | 6.09 ± 0.543 |
2.481 ± 0.324 | 4.737 ± 0.345 |
8.722 ± 0.6 | 7.519 ± 0.232 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.955 ± 0.125 | 4.662 ± 0.564 |
6.767 ± 0.686 | 6.992 ± 0.522 |
5.94 ± 0.734 | 6.316 ± 2.027 |
7.444 ± 0.793 | 2.481 ± 0.923 |
1.729 ± 0.689 | 3.759 ± 0.947 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
