
Luteibaculum oceani
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Luteibaculaceae; Luteibaculum
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
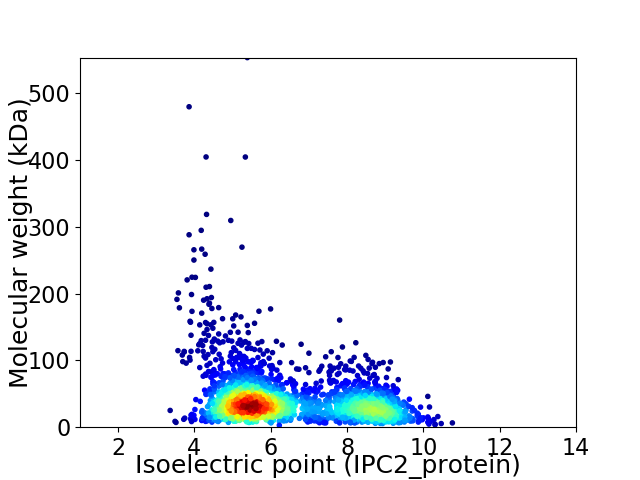
Virtual 2D-PAGE plot for 2382 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C6V129|A0A5C6V129_9FLAO Na/Pi cotransporter family protein OS=Luteibaculum oceani OX=1294296 GN=FRX97_06655 PE=4 SV=1
MM1 pKa = 7.44RR2 pKa = 11.84TLTLILLLGFLPAFLIAQNNYY23 pKa = 8.35CQPKK27 pKa = 10.15GGSPCNFQWIEE38 pKa = 4.01TFSCNNVLQNDD49 pKa = 5.91GYY51 pKa = 11.04CDD53 pKa = 4.36RR54 pKa = 11.84EE55 pKa = 4.57DD56 pKa = 4.05NDD58 pKa = 3.26EE59 pKa = 4.16TAPGFNPDD67 pKa = 3.55GYY69 pKa = 11.06SDD71 pKa = 3.84YY72 pKa = 11.45SDD74 pKa = 3.4FVVIFTPGQVTTAVLTVDD92 pKa = 3.81GNVFDD97 pKa = 5.89EE98 pKa = 4.22ISGWIDD104 pKa = 2.88WDD106 pKa = 3.55NSGTFDD112 pKa = 3.56TDD114 pKa = 3.5EE115 pKa = 5.05IITFRR120 pKa = 11.84GEE122 pKa = 4.05STVVNEE128 pKa = 3.97MTAVITVPSDD138 pKa = 3.14AVQDD142 pKa = 4.03SVILGGFRR150 pKa = 11.84IKK152 pKa = 10.41QDD154 pKa = 3.44FLEE157 pKa = 5.69PITDD161 pKa = 3.75PCADD165 pKa = 3.44IGNGEE170 pKa = 4.0IEE172 pKa = 4.8DD173 pKa = 3.7YY174 pKa = 11.49SFIVTNSGTTDD185 pKa = 3.15GGANDD190 pKa = 3.7YY191 pKa = 11.07CSARR195 pKa = 11.84GDD197 pKa = 4.06SPCNFQWIEE206 pKa = 3.88TFTCKK211 pKa = 10.39NVIQDD216 pKa = 3.91DD217 pKa = 5.55GYY219 pKa = 11.54CDD221 pKa = 5.28RR222 pKa = 11.84EE223 pKa = 4.59DD224 pKa = 4.05NDD226 pKa = 3.28EE227 pKa = 4.16TAPDD231 pKa = 4.6FDD233 pKa = 5.11PDD235 pKa = 3.98GYY237 pKa = 11.37SDD239 pKa = 4.05YY240 pKa = 11.71SDD242 pKa = 3.48FTVIFAPGEE251 pKa = 4.32VVTATLTVDD260 pKa = 3.41GNVFDD265 pKa = 5.93EE266 pKa = 4.37ISGWLDD272 pKa = 2.83WDD274 pKa = 5.0NSGTWDD280 pKa = 3.6NDD282 pKa = 3.45EE283 pKa = 4.89IITFRR288 pKa = 11.84GEE290 pKa = 4.05STVVNEE296 pKa = 3.84MTAIITVPQDD306 pKa = 3.07AVEE309 pKa = 4.39GVHH312 pKa = 6.8IIGGFRR318 pKa = 11.84IKK320 pKa = 10.36QDD322 pKa = 3.44FLEE325 pKa = 5.69PITDD329 pKa = 3.75PCADD333 pKa = 3.44IGNGEE338 pKa = 4.0IEE340 pKa = 4.79DD341 pKa = 3.71YY342 pKa = 10.92SFIVIPPDD350 pKa = 3.82NIGCASSFIPQDD362 pKa = 3.32STQNICTSTQLSWGAVSGASSYY384 pKa = 11.16QLLVLDD390 pKa = 4.79SSGDD394 pKa = 3.86TIDD397 pKa = 3.87SKK399 pKa = 11.24QVSDD403 pKa = 3.58TNLFVTGLRR412 pKa = 11.84ASEE415 pKa = 4.06AYY417 pKa = 9.18KK418 pKa = 10.26WIVKK422 pKa = 9.93PIDD425 pKa = 3.57DD426 pKa = 3.89QGRR429 pKa = 11.84KK430 pKa = 9.61SFGCDD435 pKa = 2.26TLTFFTADD443 pKa = 3.27AADD446 pKa = 3.85PTVSFSPDD454 pKa = 3.12SVDD457 pKa = 3.3VCEE460 pKa = 5.0GRR462 pKa = 11.84PLVLNPTITGGGTLSYY478 pKa = 10.37NWSGDD483 pKa = 3.38LSLLSDD489 pKa = 4.03PAVDD493 pKa = 3.35NPVFNSEE500 pKa = 3.84LPEE503 pKa = 3.91FFKK506 pKa = 11.45YY507 pKa = 10.39SLQVSNQYY515 pKa = 10.52GCSNSDD521 pKa = 3.45SLVVQVFEE529 pKa = 4.38KK530 pKa = 10.85GKK532 pKa = 10.37VVNASVNSRR541 pKa = 11.84EE542 pKa = 3.88ICFGEE547 pKa = 4.58DD548 pKa = 4.56LILTLDD554 pKa = 4.38FEE556 pKa = 6.04ADD558 pKa = 3.22SLNFLKK564 pKa = 10.63QSQNGMEE571 pKa = 4.7DD572 pKa = 3.54VSPLNQLGDD581 pKa = 3.47QYY583 pKa = 11.44IFNDD587 pKa = 3.68VDD589 pKa = 3.68TVIYY593 pKa = 10.19GVEE596 pKa = 4.23LFANGCADD604 pKa = 4.02TAWLDD609 pKa = 3.73TVVFLPQLEE618 pKa = 4.04IPEE621 pKa = 4.76IIFEE625 pKa = 4.57NPDD628 pKa = 3.41SLGPCPGEE636 pKa = 4.45DD637 pKa = 3.35YY638 pKa = 11.06LIRR641 pKa = 11.84VNNYY645 pKa = 8.64TDD647 pKa = 3.73FLSWSNGASGDD658 pKa = 4.06SIFVNEE664 pKa = 4.3TTNLTVTYY672 pKa = 10.6DD673 pKa = 3.79DD674 pKa = 4.28GCVISTDD681 pKa = 3.13TSFTFDD687 pKa = 3.74EE688 pKa = 4.71VPNTILLAIDD698 pKa = 4.27RR699 pKa = 11.84SPALLCEE706 pKa = 4.41GDD708 pKa = 4.18SIEE711 pKa = 4.11VSHH714 pKa = 7.25GIGIDD719 pKa = 3.45GFVWYY724 pKa = 10.09DD725 pKa = 3.31GDD727 pKa = 5.37DD728 pKa = 3.65SSLSKK733 pKa = 9.61WVKK736 pKa = 9.46DD737 pKa = 3.24TTLIYY742 pKa = 10.81VDD744 pKa = 5.36FISDD748 pKa = 4.02RR749 pKa = 11.84GCVKK753 pKa = 10.56VSDD756 pKa = 3.67SVQFNFKK763 pKa = 10.08PFPEE767 pKa = 4.06QAQISSLLPLDD778 pKa = 4.24SLCEE782 pKa = 3.96GDD784 pKa = 3.71VTEE787 pKa = 5.59LVSTDD792 pKa = 3.78SIYY795 pKa = 11.4DD796 pKa = 3.88LLWNTGASTPSILVSDD812 pKa = 3.94SGKK815 pKa = 10.3YY816 pKa = 8.16WLEE819 pKa = 4.07SFNGNCKK826 pKa = 9.81TISDD830 pKa = 4.18TLRR833 pKa = 11.84VDD835 pKa = 4.42FGVIPPQSSISQVVSGEE852 pKa = 4.02DD853 pKa = 3.35SLISFHH859 pKa = 7.15VADD862 pKa = 4.53HH863 pKa = 6.29YY864 pKa = 11.73DD865 pKa = 3.27WYY867 pKa = 11.12AEE869 pKa = 4.12GQLLTFDD876 pKa = 3.67EE877 pKa = 5.13RR878 pKa = 11.84IIPFVNEE885 pKa = 3.33VTYY888 pKa = 9.66TVQLRR893 pKa = 11.84SSRR896 pKa = 11.84GCLGPMSEE904 pKa = 5.03SFFKK908 pKa = 10.93NATSISEE915 pKa = 3.89FQEE918 pKa = 4.02YY919 pKa = 10.68GFVIYY924 pKa = 10.6KK925 pKa = 9.3EE926 pKa = 4.04AGRR929 pKa = 11.84SKK931 pKa = 10.9VKK933 pKa = 10.14NQRR936 pKa = 11.84PFEE939 pKa = 4.39ASLYY943 pKa = 9.3TIDD946 pKa = 4.32GRR948 pKa = 11.84LIKK951 pKa = 10.55KK952 pKa = 10.1LGSTFVFEE960 pKa = 5.14WDD962 pKa = 3.35NQGPVLLSLTLVNGKK977 pKa = 8.31QVVLKK982 pKa = 10.77VDD984 pKa = 3.31
MM1 pKa = 7.44RR2 pKa = 11.84TLTLILLLGFLPAFLIAQNNYY23 pKa = 8.35CQPKK27 pKa = 10.15GGSPCNFQWIEE38 pKa = 4.01TFSCNNVLQNDD49 pKa = 5.91GYY51 pKa = 11.04CDD53 pKa = 4.36RR54 pKa = 11.84EE55 pKa = 4.57DD56 pKa = 4.05NDD58 pKa = 3.26EE59 pKa = 4.16TAPGFNPDD67 pKa = 3.55GYY69 pKa = 11.06SDD71 pKa = 3.84YY72 pKa = 11.45SDD74 pKa = 3.4FVVIFTPGQVTTAVLTVDD92 pKa = 3.81GNVFDD97 pKa = 5.89EE98 pKa = 4.22ISGWIDD104 pKa = 2.88WDD106 pKa = 3.55NSGTFDD112 pKa = 3.56TDD114 pKa = 3.5EE115 pKa = 5.05IITFRR120 pKa = 11.84GEE122 pKa = 4.05STVVNEE128 pKa = 3.97MTAVITVPSDD138 pKa = 3.14AVQDD142 pKa = 4.03SVILGGFRR150 pKa = 11.84IKK152 pKa = 10.41QDD154 pKa = 3.44FLEE157 pKa = 5.69PITDD161 pKa = 3.75PCADD165 pKa = 3.44IGNGEE170 pKa = 4.0IEE172 pKa = 4.8DD173 pKa = 3.7YY174 pKa = 11.49SFIVTNSGTTDD185 pKa = 3.15GGANDD190 pKa = 3.7YY191 pKa = 11.07CSARR195 pKa = 11.84GDD197 pKa = 4.06SPCNFQWIEE206 pKa = 3.88TFTCKK211 pKa = 10.39NVIQDD216 pKa = 3.91DD217 pKa = 5.55GYY219 pKa = 11.54CDD221 pKa = 5.28RR222 pKa = 11.84EE223 pKa = 4.59DD224 pKa = 4.05NDD226 pKa = 3.28EE227 pKa = 4.16TAPDD231 pKa = 4.6FDD233 pKa = 5.11PDD235 pKa = 3.98GYY237 pKa = 11.37SDD239 pKa = 4.05YY240 pKa = 11.71SDD242 pKa = 3.48FTVIFAPGEE251 pKa = 4.32VVTATLTVDD260 pKa = 3.41GNVFDD265 pKa = 5.93EE266 pKa = 4.37ISGWLDD272 pKa = 2.83WDD274 pKa = 5.0NSGTWDD280 pKa = 3.6NDD282 pKa = 3.45EE283 pKa = 4.89IITFRR288 pKa = 11.84GEE290 pKa = 4.05STVVNEE296 pKa = 3.84MTAIITVPQDD306 pKa = 3.07AVEE309 pKa = 4.39GVHH312 pKa = 6.8IIGGFRR318 pKa = 11.84IKK320 pKa = 10.36QDD322 pKa = 3.44FLEE325 pKa = 5.69PITDD329 pKa = 3.75PCADD333 pKa = 3.44IGNGEE338 pKa = 4.0IEE340 pKa = 4.79DD341 pKa = 3.71YY342 pKa = 10.92SFIVIPPDD350 pKa = 3.82NIGCASSFIPQDD362 pKa = 3.32STQNICTSTQLSWGAVSGASSYY384 pKa = 11.16QLLVLDD390 pKa = 4.79SSGDD394 pKa = 3.86TIDD397 pKa = 3.87SKK399 pKa = 11.24QVSDD403 pKa = 3.58TNLFVTGLRR412 pKa = 11.84ASEE415 pKa = 4.06AYY417 pKa = 9.18KK418 pKa = 10.26WIVKK422 pKa = 9.93PIDD425 pKa = 3.57DD426 pKa = 3.89QGRR429 pKa = 11.84KK430 pKa = 9.61SFGCDD435 pKa = 2.26TLTFFTADD443 pKa = 3.27AADD446 pKa = 3.85PTVSFSPDD454 pKa = 3.12SVDD457 pKa = 3.3VCEE460 pKa = 5.0GRR462 pKa = 11.84PLVLNPTITGGGTLSYY478 pKa = 10.37NWSGDD483 pKa = 3.38LSLLSDD489 pKa = 4.03PAVDD493 pKa = 3.35NPVFNSEE500 pKa = 3.84LPEE503 pKa = 3.91FFKK506 pKa = 11.45YY507 pKa = 10.39SLQVSNQYY515 pKa = 10.52GCSNSDD521 pKa = 3.45SLVVQVFEE529 pKa = 4.38KK530 pKa = 10.85GKK532 pKa = 10.37VVNASVNSRR541 pKa = 11.84EE542 pKa = 3.88ICFGEE547 pKa = 4.58DD548 pKa = 4.56LILTLDD554 pKa = 4.38FEE556 pKa = 6.04ADD558 pKa = 3.22SLNFLKK564 pKa = 10.63QSQNGMEE571 pKa = 4.7DD572 pKa = 3.54VSPLNQLGDD581 pKa = 3.47QYY583 pKa = 11.44IFNDD587 pKa = 3.68VDD589 pKa = 3.68TVIYY593 pKa = 10.19GVEE596 pKa = 4.23LFANGCADD604 pKa = 4.02TAWLDD609 pKa = 3.73TVVFLPQLEE618 pKa = 4.04IPEE621 pKa = 4.76IIFEE625 pKa = 4.57NPDD628 pKa = 3.41SLGPCPGEE636 pKa = 4.45DD637 pKa = 3.35YY638 pKa = 11.06LIRR641 pKa = 11.84VNNYY645 pKa = 8.64TDD647 pKa = 3.73FLSWSNGASGDD658 pKa = 4.06SIFVNEE664 pKa = 4.3TTNLTVTYY672 pKa = 10.6DD673 pKa = 3.79DD674 pKa = 4.28GCVISTDD681 pKa = 3.13TSFTFDD687 pKa = 3.74EE688 pKa = 4.71VPNTILLAIDD698 pKa = 4.27RR699 pKa = 11.84SPALLCEE706 pKa = 4.41GDD708 pKa = 4.18SIEE711 pKa = 4.11VSHH714 pKa = 7.25GIGIDD719 pKa = 3.45GFVWYY724 pKa = 10.09DD725 pKa = 3.31GDD727 pKa = 5.37DD728 pKa = 3.65SSLSKK733 pKa = 9.61WVKK736 pKa = 9.46DD737 pKa = 3.24TTLIYY742 pKa = 10.81VDD744 pKa = 5.36FISDD748 pKa = 4.02RR749 pKa = 11.84GCVKK753 pKa = 10.56VSDD756 pKa = 3.67SVQFNFKK763 pKa = 10.08PFPEE767 pKa = 4.06QAQISSLLPLDD778 pKa = 4.24SLCEE782 pKa = 3.96GDD784 pKa = 3.71VTEE787 pKa = 5.59LVSTDD792 pKa = 3.78SIYY795 pKa = 11.4DD796 pKa = 3.88LLWNTGASTPSILVSDD812 pKa = 3.94SGKK815 pKa = 10.3YY816 pKa = 8.16WLEE819 pKa = 4.07SFNGNCKK826 pKa = 9.81TISDD830 pKa = 4.18TLRR833 pKa = 11.84VDD835 pKa = 4.42FGVIPPQSSISQVVSGEE852 pKa = 4.02DD853 pKa = 3.35SLISFHH859 pKa = 7.15VADD862 pKa = 4.53HH863 pKa = 6.29YY864 pKa = 11.73DD865 pKa = 3.27WYY867 pKa = 11.12AEE869 pKa = 4.12GQLLTFDD876 pKa = 3.67EE877 pKa = 5.13RR878 pKa = 11.84IIPFVNEE885 pKa = 3.33VTYY888 pKa = 9.66TVQLRR893 pKa = 11.84SSRR896 pKa = 11.84GCLGPMSEE904 pKa = 5.03SFFKK908 pKa = 10.93NATSISEE915 pKa = 3.89FQEE918 pKa = 4.02YY919 pKa = 10.68GFVIYY924 pKa = 10.6KK925 pKa = 9.3EE926 pKa = 4.04AGRR929 pKa = 11.84SKK931 pKa = 10.9VKK933 pKa = 10.14NQRR936 pKa = 11.84PFEE939 pKa = 4.39ASLYY943 pKa = 9.3TIDD946 pKa = 4.32GRR948 pKa = 11.84LIKK951 pKa = 10.55KK952 pKa = 10.1LGSTFVFEE960 pKa = 5.14WDD962 pKa = 3.35NQGPVLLSLTLVNGKK977 pKa = 8.31QVVLKK982 pKa = 10.77VDD984 pKa = 3.31
Molecular weight: 108.1 kDa
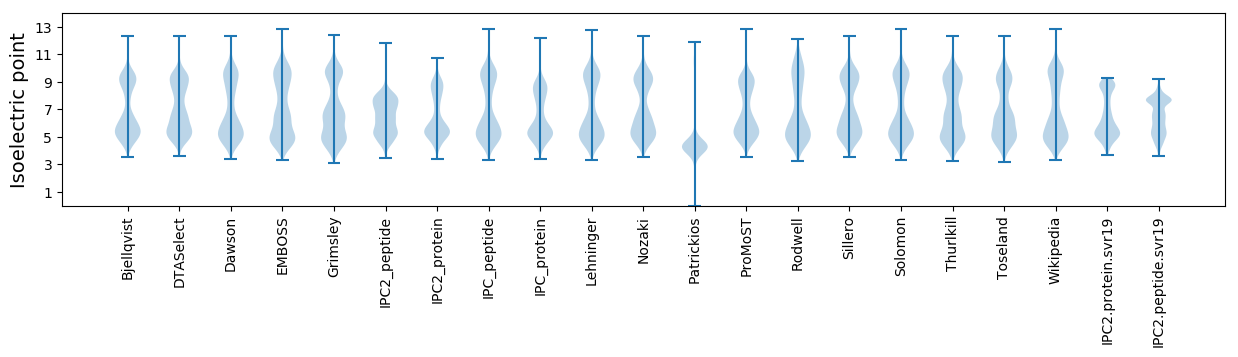
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C6V5E2|A0A5C6V5E2_9FLAO Adenosylmethionine-8-amino-7-oxononanoate aminotransferase OS=Luteibaculum oceani OX=1294296 GN=bioA PE=3 SV=1
FF1 pKa = 7.29KK2 pKa = 10.35EE3 pKa = 4.01NRR5 pKa = 11.84GKK7 pKa = 10.33IYY9 pKa = 10.78AFTRR13 pKa = 11.84LARR16 pKa = 11.84WHH18 pKa = 6.7EE19 pKa = 4.29EE20 pKa = 3.85VAQSGFKK27 pKa = 10.53SFNTISRR34 pKa = 11.84TIQNHH39 pKa = 4.44YY40 pKa = 7.68QTIINYY46 pKa = 9.29FDD48 pKa = 3.49NRR50 pKa = 11.84STNAAAEE57 pKa = 4.23SFNAKK62 pKa = 9.24IKK64 pKa = 10.71AFRR67 pKa = 11.84AQFRR71 pKa = 11.84GVRR74 pKa = 11.84KK75 pKa = 9.71IEE77 pKa = 3.94FFLFRR82 pKa = 11.84LTQIYY87 pKa = 10.32AA88 pKa = 3.37
FF1 pKa = 7.29KK2 pKa = 10.35EE3 pKa = 4.01NRR5 pKa = 11.84GKK7 pKa = 10.33IYY9 pKa = 10.78AFTRR13 pKa = 11.84LARR16 pKa = 11.84WHH18 pKa = 6.7EE19 pKa = 4.29EE20 pKa = 3.85VAQSGFKK27 pKa = 10.53SFNTISRR34 pKa = 11.84TIQNHH39 pKa = 4.44YY40 pKa = 7.68QTIINYY46 pKa = 9.29FDD48 pKa = 3.49NRR50 pKa = 11.84STNAAAEE57 pKa = 4.23SFNAKK62 pKa = 9.24IKK64 pKa = 10.71AFRR67 pKa = 11.84AQFRR71 pKa = 11.84GVRR74 pKa = 11.84KK75 pKa = 9.71IEE77 pKa = 3.94FFLFRR82 pKa = 11.84LTQIYY87 pKa = 10.32AA88 pKa = 3.37
Molecular weight: 10.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
906267 |
27 |
4889 |
380.5 |
42.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.704 ± 0.047 | 1.033 ± 0.021 |
5.545 ± 0.045 | 6.716 ± 0.058 |
5.151 ± 0.036 | 7.058 ± 0.051 |
1.755 ± 0.024 | 7.211 ± 0.039 |
7.007 ± 0.069 | 9.37 ± 0.067 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.943 ± 0.022 | 5.784 ± 0.047 |
3.94 ± 0.037 | 3.477 ± 0.028 |
3.745 ± 0.036 | 6.758 ± 0.044 |
5.381 ± 0.062 | 6.445 ± 0.045 |
1.151 ± 0.018 | 3.825 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
