
Rattus norvegicus papillomavirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Iotapapillomavirus; Iotapapillomavirus 1
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
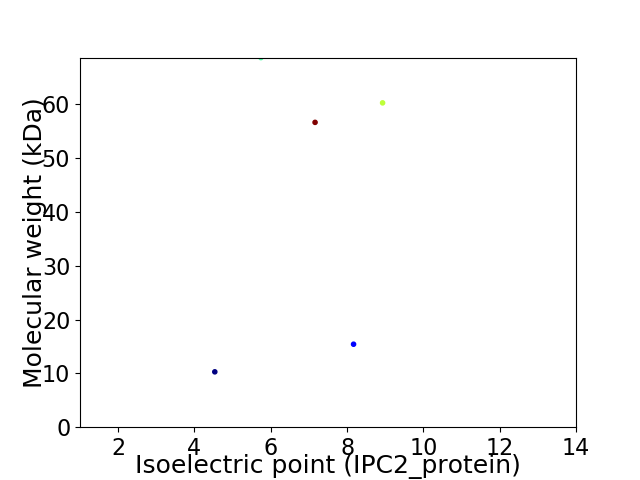
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0S2LV84|A0A0S2LV84_MNPV Replication protein E1 OS=Rattus norvegicus papillomavirus 3 OX=1756445 GN=E1 PE=3 SV=1
MM1 pKa = 7.71IGPEE5 pKa = 4.18TNRR8 pKa = 11.84TLPNDD13 pKa = 3.92PDD15 pKa = 4.3LVSLYY20 pKa = 10.26CDD22 pKa = 3.03EE23 pKa = 4.38VLEE26 pKa = 4.73DD27 pKa = 4.05EE28 pKa = 5.29PIEE31 pKa = 4.52AEE33 pKa = 4.1QSQEE37 pKa = 3.98EE38 pKa = 4.03PSIIYY43 pKa = 9.82RR44 pKa = 11.84VLIEE48 pKa = 4.66CPKK51 pKa = 9.58CDD53 pKa = 3.85RR54 pKa = 11.84IIRR57 pKa = 11.84LLCSADD63 pKa = 3.29TASIRR68 pKa = 11.84GLEE71 pKa = 3.87LLLLGGLQIICPRR84 pKa = 11.84CAEE87 pKa = 4.0KK88 pKa = 10.43HH89 pKa = 5.1GRR91 pKa = 11.84PP92 pKa = 3.78
MM1 pKa = 7.71IGPEE5 pKa = 4.18TNRR8 pKa = 11.84TLPNDD13 pKa = 3.92PDD15 pKa = 4.3LVSLYY20 pKa = 10.26CDD22 pKa = 3.03EE23 pKa = 4.38VLEE26 pKa = 4.73DD27 pKa = 4.05EE28 pKa = 5.29PIEE31 pKa = 4.52AEE33 pKa = 4.1QSQEE37 pKa = 3.98EE38 pKa = 4.03PSIIYY43 pKa = 9.82RR44 pKa = 11.84VLIEE48 pKa = 4.66CPKK51 pKa = 9.58CDD53 pKa = 3.85RR54 pKa = 11.84IIRR57 pKa = 11.84LLCSADD63 pKa = 3.29TASIRR68 pKa = 11.84GLEE71 pKa = 3.87LLLLGGLQIICPRR84 pKa = 11.84CAEE87 pKa = 4.0KK88 pKa = 10.43HH89 pKa = 5.1GRR91 pKa = 11.84PP92 pKa = 3.78
Molecular weight: 10.29 kDa
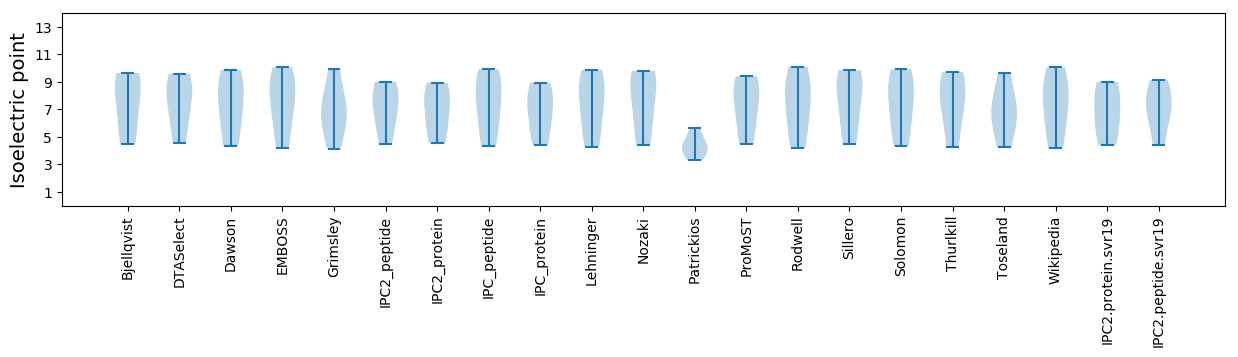
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S2LV87|A0A0S2LV87_MNPV Regulatory protein E2 OS=Rattus norvegicus papillomavirus 3 OX=1756445 GN=E2 PE=3 SV=1
MM1 pKa = 7.71EE2 pKa = 5.02SLCSRR7 pKa = 11.84LSAVQEE13 pKa = 3.99EE14 pKa = 4.78LMCIYY19 pKa = 10.74EE20 pKa = 4.28EE21 pKa = 4.55GDD23 pKa = 3.18RR24 pKa = 11.84TLATQVRR31 pKa = 11.84HH32 pKa = 4.56WQLIRR37 pKa = 11.84QEE39 pKa = 4.15QVLLHH44 pKa = 6.82AARR47 pKa = 11.84QQGHH51 pKa = 5.25TRR53 pKa = 11.84IGMQAVPSLAASQANAKK70 pKa = 9.65NAIEE74 pKa = 3.98MHH76 pKa = 6.77LLLQSLLQSPYY87 pKa = 11.13AKK89 pKa = 10.42EE90 pKa = 3.66PWTLSEE96 pKa = 4.13TSRR99 pKa = 11.84DD100 pKa = 3.64LYY102 pKa = 10.82LAPPSHH108 pKa = 6.09TLKK111 pKa = 10.86KK112 pKa = 10.05SGQTIEE118 pKa = 3.99VLFDD122 pKa = 3.77GDD124 pKa = 4.53EE125 pKa = 4.33NNSMQYY131 pKa = 9.58TKK133 pKa = 9.95WLQVFYY139 pKa = 10.79TDD141 pKa = 5.76DD142 pKa = 3.51RR143 pKa = 11.84GMWKK147 pKa = 9.99RR148 pKa = 11.84VPSVCDD154 pKa = 3.08ATGIFFDD161 pKa = 4.26RR162 pKa = 11.84NGHH165 pKa = 5.09RR166 pKa = 11.84EE167 pKa = 3.86YY168 pKa = 10.97YY169 pKa = 10.51VRR171 pKa = 11.84FEE173 pKa = 4.11KK174 pKa = 10.72EE175 pKa = 3.42AMRR178 pKa = 11.84FSVTGTWQAVDD189 pKa = 3.16EE190 pKa = 4.49GKK192 pKa = 10.19IYY194 pKa = 10.82SSVVPVASSTPTTGPPRR211 pKa = 11.84GPAVHH216 pKa = 7.29PDD218 pKa = 3.35GVSATGHH225 pKa = 6.16TSPNWDD231 pKa = 3.92CSRR234 pKa = 11.84HH235 pKa = 5.51SPLPSGGGEE244 pKa = 3.84DD245 pKa = 3.12SGEE248 pKa = 4.03EE249 pKa = 4.22EE250 pKa = 4.44KK251 pKa = 11.25VHH253 pKa = 5.74TRR255 pKa = 11.84PEE257 pKa = 4.37YY258 pKa = 10.12TDD260 pKa = 3.32HH261 pKa = 7.08SYY263 pKa = 11.19NRR265 pKa = 11.84QTSPPSFRR273 pKa = 11.84GEE275 pKa = 3.67QSVGGEE281 pKa = 3.85QHH283 pKa = 6.71RR284 pKa = 11.84EE285 pKa = 3.49RR286 pKa = 11.84ARR288 pKa = 11.84GLGVPAGDD296 pKa = 3.42VPEE299 pKa = 4.39VPRR302 pKa = 11.84AADD305 pKa = 3.71KK306 pKa = 7.79EE307 pKa = 4.86TKK309 pKa = 10.39SPEE312 pKa = 4.69PISKK316 pKa = 10.16RR317 pKa = 11.84GPGRR321 pKa = 11.84PRR323 pKa = 11.84KK324 pKa = 9.49GPSAAGEE331 pKa = 4.27TTTDD335 pKa = 3.37GEE337 pKa = 4.36PRR339 pKa = 11.84SKK341 pKa = 10.42KK342 pKa = 9.88RR343 pKa = 11.84PRR345 pKa = 11.84DD346 pKa = 3.27RR347 pKa = 11.84TGGYY351 pKa = 7.46RR352 pKa = 11.84TRR354 pKa = 11.84KK355 pKa = 8.91RR356 pKa = 11.84RR357 pKa = 11.84RR358 pKa = 11.84TKK360 pKa = 10.2RR361 pKa = 11.84WATPPRR367 pKa = 11.84EE368 pKa = 4.04RR369 pKa = 11.84GPDD372 pKa = 3.48HH373 pKa = 7.4VDD375 pKa = 3.69LNATNKK381 pKa = 10.32NEE383 pKa = 3.71GRR385 pKa = 11.84PNGPVNCSPPTPNPPDD401 pKa = 3.8LSAIPPTPGVRR412 pKa = 11.84VAGQSTPAGTSRR424 pKa = 11.84APPTLVAEE432 pKa = 4.69PDD434 pKa = 3.35AGRR437 pKa = 11.84TPAAAAAGLRR447 pKa = 11.84SATPGSTARR456 pKa = 11.84IRR458 pKa = 11.84RR459 pKa = 11.84PRR461 pKa = 11.84VGPGPPCLLIIKK473 pKa = 7.66GTPNGVKK480 pKa = 10.26CLRR483 pKa = 11.84YY484 pKa = 9.41RR485 pKa = 11.84LKK487 pKa = 9.26TQHH490 pKa = 7.0PSTFQFVSTTWQWVPATGSSRR511 pKa = 11.84VGRR514 pKa = 11.84ARR516 pKa = 11.84ILVMCSDD523 pKa = 3.33QTQRR527 pKa = 11.84NIFLDD532 pKa = 3.89TVKK535 pKa = 10.22IPAGMTVEE543 pKa = 4.2QCSMLAII550 pKa = 4.49
MM1 pKa = 7.71EE2 pKa = 5.02SLCSRR7 pKa = 11.84LSAVQEE13 pKa = 3.99EE14 pKa = 4.78LMCIYY19 pKa = 10.74EE20 pKa = 4.28EE21 pKa = 4.55GDD23 pKa = 3.18RR24 pKa = 11.84TLATQVRR31 pKa = 11.84HH32 pKa = 4.56WQLIRR37 pKa = 11.84QEE39 pKa = 4.15QVLLHH44 pKa = 6.82AARR47 pKa = 11.84QQGHH51 pKa = 5.25TRR53 pKa = 11.84IGMQAVPSLAASQANAKK70 pKa = 9.65NAIEE74 pKa = 3.98MHH76 pKa = 6.77LLLQSLLQSPYY87 pKa = 11.13AKK89 pKa = 10.42EE90 pKa = 3.66PWTLSEE96 pKa = 4.13TSRR99 pKa = 11.84DD100 pKa = 3.64LYY102 pKa = 10.82LAPPSHH108 pKa = 6.09TLKK111 pKa = 10.86KK112 pKa = 10.05SGQTIEE118 pKa = 3.99VLFDD122 pKa = 3.77GDD124 pKa = 4.53EE125 pKa = 4.33NNSMQYY131 pKa = 9.58TKK133 pKa = 9.95WLQVFYY139 pKa = 10.79TDD141 pKa = 5.76DD142 pKa = 3.51RR143 pKa = 11.84GMWKK147 pKa = 9.99RR148 pKa = 11.84VPSVCDD154 pKa = 3.08ATGIFFDD161 pKa = 4.26RR162 pKa = 11.84NGHH165 pKa = 5.09RR166 pKa = 11.84EE167 pKa = 3.86YY168 pKa = 10.97YY169 pKa = 10.51VRR171 pKa = 11.84FEE173 pKa = 4.11KK174 pKa = 10.72EE175 pKa = 3.42AMRR178 pKa = 11.84FSVTGTWQAVDD189 pKa = 3.16EE190 pKa = 4.49GKK192 pKa = 10.19IYY194 pKa = 10.82SSVVPVASSTPTTGPPRR211 pKa = 11.84GPAVHH216 pKa = 7.29PDD218 pKa = 3.35GVSATGHH225 pKa = 6.16TSPNWDD231 pKa = 3.92CSRR234 pKa = 11.84HH235 pKa = 5.51SPLPSGGGEE244 pKa = 3.84DD245 pKa = 3.12SGEE248 pKa = 4.03EE249 pKa = 4.22EE250 pKa = 4.44KK251 pKa = 11.25VHH253 pKa = 5.74TRR255 pKa = 11.84PEE257 pKa = 4.37YY258 pKa = 10.12TDD260 pKa = 3.32HH261 pKa = 7.08SYY263 pKa = 11.19NRR265 pKa = 11.84QTSPPSFRR273 pKa = 11.84GEE275 pKa = 3.67QSVGGEE281 pKa = 3.85QHH283 pKa = 6.71RR284 pKa = 11.84EE285 pKa = 3.49RR286 pKa = 11.84ARR288 pKa = 11.84GLGVPAGDD296 pKa = 3.42VPEE299 pKa = 4.39VPRR302 pKa = 11.84AADD305 pKa = 3.71KK306 pKa = 7.79EE307 pKa = 4.86TKK309 pKa = 10.39SPEE312 pKa = 4.69PISKK316 pKa = 10.16RR317 pKa = 11.84GPGRR321 pKa = 11.84PRR323 pKa = 11.84KK324 pKa = 9.49GPSAAGEE331 pKa = 4.27TTTDD335 pKa = 3.37GEE337 pKa = 4.36PRR339 pKa = 11.84SKK341 pKa = 10.42KK342 pKa = 9.88RR343 pKa = 11.84PRR345 pKa = 11.84DD346 pKa = 3.27RR347 pKa = 11.84TGGYY351 pKa = 7.46RR352 pKa = 11.84TRR354 pKa = 11.84KK355 pKa = 8.91RR356 pKa = 11.84RR357 pKa = 11.84RR358 pKa = 11.84TKK360 pKa = 10.2RR361 pKa = 11.84WATPPRR367 pKa = 11.84EE368 pKa = 4.04RR369 pKa = 11.84GPDD372 pKa = 3.48HH373 pKa = 7.4VDD375 pKa = 3.69LNATNKK381 pKa = 10.32NEE383 pKa = 3.71GRR385 pKa = 11.84PNGPVNCSPPTPNPPDD401 pKa = 3.8LSAIPPTPGVRR412 pKa = 11.84VAGQSTPAGTSRR424 pKa = 11.84APPTLVAEE432 pKa = 4.69PDD434 pKa = 3.35AGRR437 pKa = 11.84TPAAAAAGLRR447 pKa = 11.84SATPGSTARR456 pKa = 11.84IRR458 pKa = 11.84RR459 pKa = 11.84PRR461 pKa = 11.84VGPGPPCLLIIKK473 pKa = 7.66GTPNGVKK480 pKa = 10.26CLRR483 pKa = 11.84YY484 pKa = 9.41RR485 pKa = 11.84LKK487 pKa = 9.26TQHH490 pKa = 7.0PSTFQFVSTTWQWVPATGSSRR511 pKa = 11.84VGRR514 pKa = 11.84ARR516 pKa = 11.84ILVMCSDD523 pKa = 3.33QTQRR527 pKa = 11.84NIFLDD532 pKa = 3.89TVKK535 pKa = 10.22IPAGMTVEE543 pKa = 4.2QCSMLAII550 pKa = 4.49
Molecular weight: 60.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1876 |
92 |
600 |
375.2 |
42.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.237 ± 0.716 | 2.719 ± 0.662 |
5.384 ± 0.509 | 6.397 ± 0.445 |
3.625 ± 0.711 | 6.716 ± 0.72 |
1.972 ± 0.186 | 3.945 ± 0.55 |
4.797 ± 0.498 | 8.102 ± 0.844 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.972 ± 0.308 | 4.211 ± 0.645 |
6.61 ± 1.228 | 4.478 ± 0.218 |
7.729 ± 0.921 | 7.409 ± 0.3 |
6.61 ± 0.566 | 6.183 ± 0.442 |
1.652 ± 0.208 | 3.252 ± 0.56 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
