
Tupaia paramyxovirus (TPMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Narmovirus; Tupaia narmovirus
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
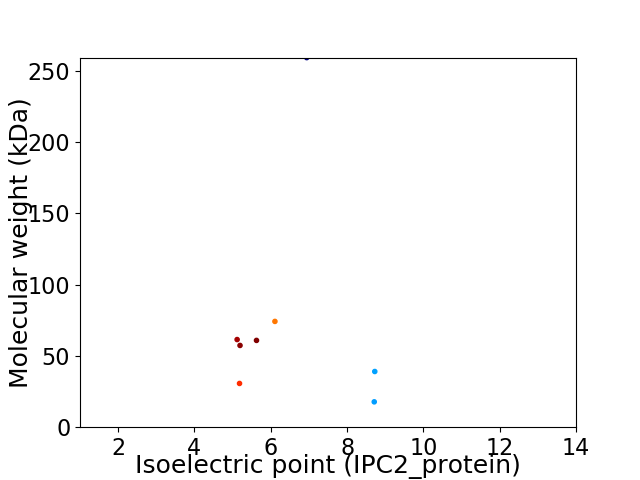
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9WS40|NCAP_TPMV Nucleoprotein OS=Tupaia paramyxovirus OX=92129 GN=N PE=3 SV=1
MM1 pKa = 7.91ADD3 pKa = 4.19LFSKK7 pKa = 11.11VNDD10 pKa = 3.71FQKK13 pKa = 10.86YY14 pKa = 5.08RR15 pKa = 11.84TNLGRR20 pKa = 11.84QGGLTVKK27 pKa = 10.5LVGVRR32 pKa = 11.84STVVVLVPSTKK43 pKa = 9.96DD44 pKa = 2.9HH45 pKa = 6.45RR46 pKa = 11.84LRR48 pKa = 11.84WKK50 pKa = 10.66LIRR53 pKa = 11.84LLTLAVYY60 pKa = 10.55NDD62 pKa = 4.24SLPDD66 pKa = 4.1SISIGALLSLLAISFEE82 pKa = 4.03QPAAVIRR89 pKa = 11.84GLLSDD94 pKa = 4.94PDD96 pKa = 4.93LEE98 pKa = 4.34VQMIEE103 pKa = 5.1VSLDD107 pKa = 3.31DD108 pKa = 3.34QGEE111 pKa = 3.97IRR113 pKa = 11.84FAARR117 pKa = 11.84GDD119 pKa = 3.07ILTRR123 pKa = 11.84YY124 pKa = 9.1KK125 pKa = 10.57DD126 pKa = 3.34AYY128 pKa = 9.96FEE130 pKa = 4.75KK131 pKa = 10.51IRR133 pKa = 11.84DD134 pKa = 4.01FPNPDD139 pKa = 3.32DD140 pKa = 4.77DD141 pKa = 5.05LAIFEE146 pKa = 4.77DD147 pKa = 4.9PEE149 pKa = 5.54LGDD152 pKa = 4.02YY153 pKa = 11.32SDD155 pKa = 3.68ITQDD159 pKa = 3.41EE160 pKa = 4.64YY161 pKa = 11.62QAMITTITIQLWILLTKK178 pKa = 10.58AVTAPDD184 pKa = 3.73TAHH187 pKa = 7.47DD188 pKa = 3.81SEE190 pKa = 3.96QRR192 pKa = 11.84RR193 pKa = 11.84FIKK196 pKa = 10.49YY197 pKa = 8.08LQQRR201 pKa = 11.84KK202 pKa = 9.35AYY204 pKa = 9.8AAFKK208 pKa = 9.08FTTIFTEE215 pKa = 4.14RR216 pKa = 11.84VRR218 pKa = 11.84RR219 pKa = 11.84KK220 pKa = 9.03IAQSLSIRR228 pKa = 11.84KK229 pKa = 9.04FMVSIMLEE237 pKa = 4.04VRR239 pKa = 11.84KK240 pKa = 10.47SGSAKK245 pKa = 10.18GRR247 pKa = 11.84ISEE250 pKa = 4.77CIADD254 pKa = 3.43VSAYY258 pKa = 9.62IEE260 pKa = 4.06EE261 pKa = 4.78AGLSGFILTLKK272 pKa = 10.32YY273 pKa = 10.77GIGTRR278 pKa = 11.84FPVLALNAFQSDD290 pKa = 3.5LSVIRR295 pKa = 11.84NLIDD299 pKa = 3.71LYY301 pKa = 11.45KK302 pKa = 11.02SMGTIAPFMVLIEE315 pKa = 5.1DD316 pKa = 3.8ATQVKK321 pKa = 8.69FAPGNYY327 pKa = 9.22SLLWSFAMGVGTALDD342 pKa = 3.32HH343 pKa = 7.49AMNNLNINRR352 pKa = 11.84DD353 pKa = 3.86YY354 pKa = 11.57LEE356 pKa = 4.02PSYY359 pKa = 10.88FRR361 pKa = 11.84LGQEE365 pKa = 4.03VVRR368 pKa = 11.84LSEE371 pKa = 4.32STVDD375 pKa = 3.21RR376 pKa = 11.84SMAQEE381 pKa = 4.22LGIDD385 pKa = 3.79PTSEE389 pKa = 4.0DD390 pKa = 3.79LIMRR394 pKa = 11.84AVQAAGVGSRR404 pKa = 11.84DD405 pKa = 2.95PDD407 pKa = 3.33AARR410 pKa = 11.84RR411 pKa = 11.84TGRR414 pKa = 11.84FQVADD419 pKa = 3.52IQIDD423 pKa = 3.72EE424 pKa = 4.84GPVDD428 pKa = 4.22LATEE432 pKa = 4.75AEE434 pKa = 4.26DD435 pKa = 3.77QTTKK439 pKa = 11.15DD440 pKa = 2.97NEE442 pKa = 4.02QRR444 pKa = 11.84IKK446 pKa = 11.19VPDD449 pKa = 3.66PRR451 pKa = 11.84GSIGQSNAQFQPPKK465 pKa = 8.08PQLRR469 pKa = 11.84GRR471 pKa = 11.84VMPPEE476 pKa = 4.63RR477 pKa = 11.84KK478 pKa = 8.42PTDD481 pKa = 3.46QQKK484 pKa = 10.15NLQDD488 pKa = 3.39QRR490 pKa = 11.84PRR492 pKa = 11.84PSATPRR498 pKa = 11.84RR499 pKa = 11.84LTKK502 pKa = 10.53DD503 pKa = 2.92AEE505 pKa = 4.49DD506 pKa = 5.15NIDD509 pKa = 3.87QLFAQYY515 pKa = 10.87DD516 pKa = 3.85SGVAAPEE523 pKa = 4.09DD524 pKa = 3.86VTLVTSDD531 pKa = 3.24STSPARR537 pKa = 11.84SSGTGSDD544 pKa = 3.15MDD546 pKa = 6.19LINQSPP552 pKa = 3.25
MM1 pKa = 7.91ADD3 pKa = 4.19LFSKK7 pKa = 11.11VNDD10 pKa = 3.71FQKK13 pKa = 10.86YY14 pKa = 5.08RR15 pKa = 11.84TNLGRR20 pKa = 11.84QGGLTVKK27 pKa = 10.5LVGVRR32 pKa = 11.84STVVVLVPSTKK43 pKa = 9.96DD44 pKa = 2.9HH45 pKa = 6.45RR46 pKa = 11.84LRR48 pKa = 11.84WKK50 pKa = 10.66LIRR53 pKa = 11.84LLTLAVYY60 pKa = 10.55NDD62 pKa = 4.24SLPDD66 pKa = 4.1SISIGALLSLLAISFEE82 pKa = 4.03QPAAVIRR89 pKa = 11.84GLLSDD94 pKa = 4.94PDD96 pKa = 4.93LEE98 pKa = 4.34VQMIEE103 pKa = 5.1VSLDD107 pKa = 3.31DD108 pKa = 3.34QGEE111 pKa = 3.97IRR113 pKa = 11.84FAARR117 pKa = 11.84GDD119 pKa = 3.07ILTRR123 pKa = 11.84YY124 pKa = 9.1KK125 pKa = 10.57DD126 pKa = 3.34AYY128 pKa = 9.96FEE130 pKa = 4.75KK131 pKa = 10.51IRR133 pKa = 11.84DD134 pKa = 4.01FPNPDD139 pKa = 3.32DD140 pKa = 4.77DD141 pKa = 5.05LAIFEE146 pKa = 4.77DD147 pKa = 4.9PEE149 pKa = 5.54LGDD152 pKa = 4.02YY153 pKa = 11.32SDD155 pKa = 3.68ITQDD159 pKa = 3.41EE160 pKa = 4.64YY161 pKa = 11.62QAMITTITIQLWILLTKK178 pKa = 10.58AVTAPDD184 pKa = 3.73TAHH187 pKa = 7.47DD188 pKa = 3.81SEE190 pKa = 3.96QRR192 pKa = 11.84RR193 pKa = 11.84FIKK196 pKa = 10.49YY197 pKa = 8.08LQQRR201 pKa = 11.84KK202 pKa = 9.35AYY204 pKa = 9.8AAFKK208 pKa = 9.08FTTIFTEE215 pKa = 4.14RR216 pKa = 11.84VRR218 pKa = 11.84RR219 pKa = 11.84KK220 pKa = 9.03IAQSLSIRR228 pKa = 11.84KK229 pKa = 9.04FMVSIMLEE237 pKa = 4.04VRR239 pKa = 11.84KK240 pKa = 10.47SGSAKK245 pKa = 10.18GRR247 pKa = 11.84ISEE250 pKa = 4.77CIADD254 pKa = 3.43VSAYY258 pKa = 9.62IEE260 pKa = 4.06EE261 pKa = 4.78AGLSGFILTLKK272 pKa = 10.32YY273 pKa = 10.77GIGTRR278 pKa = 11.84FPVLALNAFQSDD290 pKa = 3.5LSVIRR295 pKa = 11.84NLIDD299 pKa = 3.71LYY301 pKa = 11.45KK302 pKa = 11.02SMGTIAPFMVLIEE315 pKa = 5.1DD316 pKa = 3.8ATQVKK321 pKa = 8.69FAPGNYY327 pKa = 9.22SLLWSFAMGVGTALDD342 pKa = 3.32HH343 pKa = 7.49AMNNLNINRR352 pKa = 11.84DD353 pKa = 3.86YY354 pKa = 11.57LEE356 pKa = 4.02PSYY359 pKa = 10.88FRR361 pKa = 11.84LGQEE365 pKa = 4.03VVRR368 pKa = 11.84LSEE371 pKa = 4.32STVDD375 pKa = 3.21RR376 pKa = 11.84SMAQEE381 pKa = 4.22LGIDD385 pKa = 3.79PTSEE389 pKa = 4.0DD390 pKa = 3.79LIMRR394 pKa = 11.84AVQAAGVGSRR404 pKa = 11.84DD405 pKa = 2.95PDD407 pKa = 3.33AARR410 pKa = 11.84RR411 pKa = 11.84TGRR414 pKa = 11.84FQVADD419 pKa = 3.52IQIDD423 pKa = 3.72EE424 pKa = 4.84GPVDD428 pKa = 4.22LATEE432 pKa = 4.75AEE434 pKa = 4.26DD435 pKa = 3.77QTTKK439 pKa = 11.15DD440 pKa = 2.97NEE442 pKa = 4.02QRR444 pKa = 11.84IKK446 pKa = 11.19VPDD449 pKa = 3.66PRR451 pKa = 11.84GSIGQSNAQFQPPKK465 pKa = 8.08PQLRR469 pKa = 11.84GRR471 pKa = 11.84VMPPEE476 pKa = 4.63RR477 pKa = 11.84KK478 pKa = 8.42PTDD481 pKa = 3.46QQKK484 pKa = 10.15NLQDD488 pKa = 3.39QRR490 pKa = 11.84PRR492 pKa = 11.84PSATPRR498 pKa = 11.84RR499 pKa = 11.84LTKK502 pKa = 10.53DD503 pKa = 2.92AEE505 pKa = 4.49DD506 pKa = 5.15NIDD509 pKa = 3.87QLFAQYY515 pKa = 10.87DD516 pKa = 3.85SGVAAPEE523 pKa = 4.09DD524 pKa = 3.86VTLVTSDD531 pKa = 3.24STSPARR537 pKa = 11.84SSGTGSDD544 pKa = 3.15MDD546 pKa = 6.19LINQSPP552 pKa = 3.25
Molecular weight: 61.52 kDa
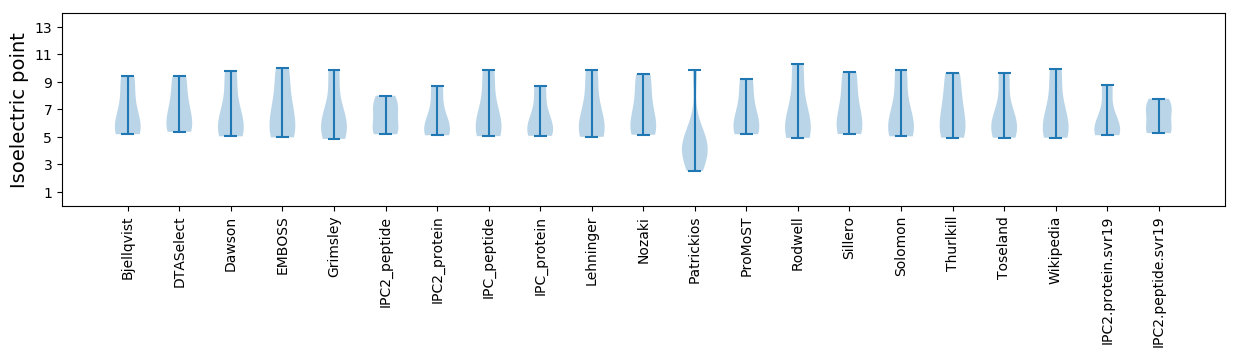
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9QM81|V_TPMV Non-structural protein V OS=Tupaia paramyxovirus OX=92129 GN=P/V PE=3 SV=1
MM1 pKa = 7.13YY2 pKa = 9.38PSAPRR7 pKa = 11.84IDD9 pKa = 3.82EE10 pKa = 4.4APVVDD15 pKa = 5.14CEE17 pKa = 4.56YY18 pKa = 11.14EE19 pKa = 4.4FIPTTWLEE27 pKa = 3.88KK28 pKa = 10.7GYY30 pKa = 10.47LSAMKK35 pKa = 10.5VEE37 pKa = 4.53SDD39 pKa = 3.66HH40 pKa = 6.38NGKK43 pKa = 9.97IIPSVRR49 pKa = 11.84VVNPGWGEE57 pKa = 4.13RR58 pKa = 11.84KK59 pKa = 8.13TSGYY63 pKa = 9.47MYY65 pKa = 10.58LIMHH69 pKa = 7.25GIVEE73 pKa = 4.72DD74 pKa = 4.01VPKK77 pKa = 11.09DD78 pKa = 3.05GDD80 pKa = 3.58TEE82 pKa = 3.95QRR84 pKa = 11.84YY85 pKa = 9.69SGKK88 pKa = 8.96TYY90 pKa = 10.51AAFPLGVGKK99 pKa = 10.96SNATPDD105 pKa = 4.32DD106 pKa = 4.72LLTSMNKK113 pKa = 9.83LQITVRR119 pKa = 11.84RR120 pKa = 11.84TAGAGEE126 pKa = 4.21RR127 pKa = 11.84IVFGNNAPLGALFPWRR143 pKa = 11.84RR144 pKa = 11.84VLDD147 pKa = 4.58FGAVFTAYY155 pKa = 9.32KK156 pKa = 9.95VCLSVEE162 pKa = 4.58SISLFTPQRR171 pKa = 11.84FRR173 pKa = 11.84PLFLTVTLLTDD184 pKa = 3.28NGLYY188 pKa = 10.15KK189 pKa = 10.67APSLFADD196 pKa = 5.05FRR198 pKa = 11.84ASKK201 pKa = 10.5AVSFNLLARR210 pKa = 11.84LTVNNKK216 pKa = 9.56SGKK219 pKa = 9.93DD220 pKa = 3.68YY221 pKa = 11.06LATAPASDD229 pKa = 3.68TKK231 pKa = 10.87QVVSFMVHH239 pKa = 5.6IGNFVRR245 pKa = 11.84KK246 pKa = 10.13GGDD249 pKa = 3.19VYY251 pKa = 11.67SNSYY255 pKa = 9.47CKK257 pKa = 10.49KK258 pKa = 10.39KK259 pKa = 9.74IDD261 pKa = 4.57RR262 pKa = 11.84MDD264 pKa = 3.34LQFALGAVGGLSFHH278 pKa = 6.86IKK280 pKa = 10.3INGKK284 pKa = 7.38MSKK287 pKa = 8.46TLMTQLGFHH296 pKa = 7.11RR297 pKa = 11.84NLCYY301 pKa = 10.62SIMDD305 pKa = 4.33INPDD309 pKa = 3.55LNKK312 pKa = 10.56KK313 pKa = 8.53IWNSSCRR320 pKa = 11.84ITSVAAILQPSVSKK334 pKa = 10.75DD335 pKa = 3.11FKK337 pKa = 10.37IYY339 pKa = 10.54HH340 pKa = 6.85DD341 pKa = 3.96VFIDD345 pKa = 3.53NTGKK349 pKa = 10.5IMGG352 pKa = 4.27
MM1 pKa = 7.13YY2 pKa = 9.38PSAPRR7 pKa = 11.84IDD9 pKa = 3.82EE10 pKa = 4.4APVVDD15 pKa = 5.14CEE17 pKa = 4.56YY18 pKa = 11.14EE19 pKa = 4.4FIPTTWLEE27 pKa = 3.88KK28 pKa = 10.7GYY30 pKa = 10.47LSAMKK35 pKa = 10.5VEE37 pKa = 4.53SDD39 pKa = 3.66HH40 pKa = 6.38NGKK43 pKa = 9.97IIPSVRR49 pKa = 11.84VVNPGWGEE57 pKa = 4.13RR58 pKa = 11.84KK59 pKa = 8.13TSGYY63 pKa = 9.47MYY65 pKa = 10.58LIMHH69 pKa = 7.25GIVEE73 pKa = 4.72DD74 pKa = 4.01VPKK77 pKa = 11.09DD78 pKa = 3.05GDD80 pKa = 3.58TEE82 pKa = 3.95QRR84 pKa = 11.84YY85 pKa = 9.69SGKK88 pKa = 8.96TYY90 pKa = 10.51AAFPLGVGKK99 pKa = 10.96SNATPDD105 pKa = 4.32DD106 pKa = 4.72LLTSMNKK113 pKa = 9.83LQITVRR119 pKa = 11.84RR120 pKa = 11.84TAGAGEE126 pKa = 4.21RR127 pKa = 11.84IVFGNNAPLGALFPWRR143 pKa = 11.84RR144 pKa = 11.84VLDD147 pKa = 4.58FGAVFTAYY155 pKa = 9.32KK156 pKa = 9.95VCLSVEE162 pKa = 4.58SISLFTPQRR171 pKa = 11.84FRR173 pKa = 11.84PLFLTVTLLTDD184 pKa = 3.28NGLYY188 pKa = 10.15KK189 pKa = 10.67APSLFADD196 pKa = 5.05FRR198 pKa = 11.84ASKK201 pKa = 10.5AVSFNLLARR210 pKa = 11.84LTVNNKK216 pKa = 9.56SGKK219 pKa = 9.93DD220 pKa = 3.68YY221 pKa = 11.06LATAPASDD229 pKa = 3.68TKK231 pKa = 10.87QVVSFMVHH239 pKa = 5.6IGNFVRR245 pKa = 11.84KK246 pKa = 10.13GGDD249 pKa = 3.19VYY251 pKa = 11.67SNSYY255 pKa = 9.47CKK257 pKa = 10.49KK258 pKa = 10.39KK259 pKa = 9.74IDD261 pKa = 4.57RR262 pKa = 11.84MDD264 pKa = 3.34LQFALGAVGGLSFHH278 pKa = 6.86IKK280 pKa = 10.3INGKK284 pKa = 7.38MSKK287 pKa = 8.46TLMTQLGFHH296 pKa = 7.11RR297 pKa = 11.84NLCYY301 pKa = 10.62SIMDD305 pKa = 4.33INPDD309 pKa = 3.55LNKK312 pKa = 10.56KK313 pKa = 8.53IWNSSCRR320 pKa = 11.84ITSVAAILQPSVSKK334 pKa = 10.75DD335 pKa = 3.11FKK337 pKa = 10.37IYY339 pKa = 10.54HH340 pKa = 6.85DD341 pKa = 3.96VFIDD345 pKa = 3.53NTGKK349 pKa = 10.5IMGG352 pKa = 4.27
Molecular weight: 39.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5354 |
153 |
2270 |
669.3 |
75.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.753 ± 0.671 | 1.886 ± 0.303 |
5.977 ± 0.394 | 5.454 ± 0.699 |
3.044 ± 0.492 | 5.902 ± 0.508 |
2.26 ± 0.234 | 6.911 ± 0.533 |
5.902 ± 0.344 | 10.105 ± 0.554 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.409 ± 0.234 | 5.118 ± 0.46 |
5.043 ± 0.404 | 4.053 ± 0.414 |
4.968 ± 0.485 | 8.797 ± 0.413 |
6.22 ± 0.392 | 4.968 ± 0.366 |
1.102 ± 0.164 | 4.128 ± 0.816 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
