
Nitrolancea hollandica Lb
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Thermomicrobia; Sphaerobacteridae; Sphaerobacterales; Sphaerobacterineae; Sphaerobacteraceae; Nitrolancea; Nitrolancea hollandica
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
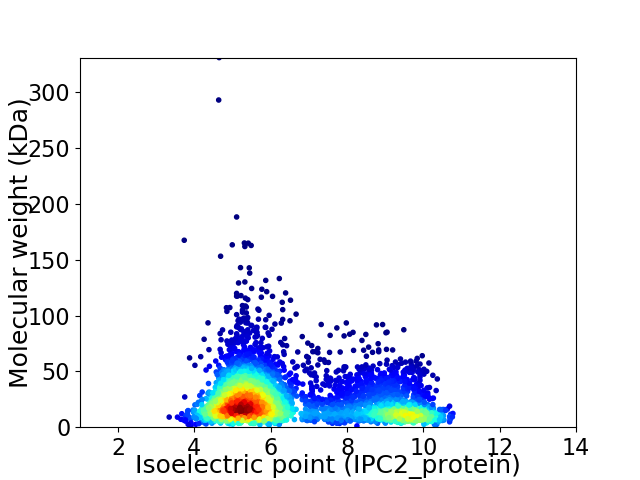
Virtual 2D-PAGE plot for 3969 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I4EJR4|I4EJR4_9CHLR Putative ABC-type branched-chain amino acid transport systems periplasmic binding component OS=Nitrolancea hollandica Lb OX=1129897 GN=NITHO_4210003 PE=4 SV=1
MM1 pKa = 7.71RR2 pKa = 11.84RR3 pKa = 11.84VLWAIGLTLGLLSSLLFAGTAMAAGLGSVGTLSGAVSGVTDD44 pKa = 3.9TVSSVADD51 pKa = 3.56PVTATVSDD59 pKa = 3.85VAGAISDD66 pKa = 3.85AGSNGAGNNVNSGNTTSAGLIAVNDD91 pKa = 4.09LVDD94 pKa = 3.85VSALNGGNGGDD105 pKa = 3.3AGLNVSSPDD114 pKa = 3.34VTVGGLVAVNDD125 pKa = 4.03AVDD128 pKa = 3.46VSLLNGGNGDD138 pKa = 3.76GSGAGNNTNTGNTTTAGLIAVNDD161 pKa = 4.14LADD164 pKa = 3.47VSALNGANANGSHH177 pKa = 6.65AGSNAGNNTNGDD189 pKa = 3.67NTTSAGLIAVNDD201 pKa = 4.33GVDD204 pKa = 3.15ASLLNGANANGSHH217 pKa = 6.2SEE219 pKa = 3.99ANAGNNTNSGNTTSAGLVAVNDD241 pKa = 5.5GIDD244 pKa = 3.16ASLLNGANANGNHH257 pKa = 6.49AEE259 pKa = 4.1ANAGNNANSGNVTSGGLAGINDD281 pKa = 5.43GIDD284 pKa = 2.9ASFGNGANANGSLSGANAGNNVNTGNTTSSGVAGVNDD321 pKa = 5.59GIDD324 pKa = 3.2AGLLNGGNANGKK336 pKa = 8.24HH337 pKa = 5.77AEE339 pKa = 4.16VNAGNNVNTGNTTTGGLVGANDD361 pKa = 5.53LIDD364 pKa = 3.58ATLINGANANGDD376 pKa = 3.88HH377 pKa = 6.29AHH379 pKa = 7.01ANAGNNVNAGNTTSGGLIGINDD401 pKa = 4.46LLGATALNGANANGEE416 pKa = 4.17GARR419 pKa = 11.84ANAGNNINSGNITSGGLIAINDD441 pKa = 4.31PLDD444 pKa = 3.93LSLLNGANADD454 pKa = 3.59GSVGNHH460 pKa = 5.92GCTQNCGGDD469 pKa = 3.38NGCEE473 pKa = 3.9HH474 pKa = 7.79DD475 pKa = 4.09CGGDD479 pKa = 3.21HH480 pKa = 7.36GCGHH484 pKa = 7.57DD485 pKa = 3.51CGGDD489 pKa = 3.16HH490 pKa = 7.23GCTHH494 pKa = 6.61NCGGDD499 pKa = 3.64HH500 pKa = 6.46EE501 pKa = 5.28CSSNCGGNHH510 pKa = 7.04GGCTSNCSGNDD521 pKa = 3.27GHH523 pKa = 7.28CIANCEE529 pKa = 4.16GTSDD533 pKa = 3.49GGKK536 pKa = 10.09NNHH539 pKa = 6.54EE540 pKa = 4.47GNDD543 pKa = 4.13PIDD546 pKa = 4.16GGNHH550 pKa = 6.47NGGDD554 pKa = 3.84DD555 pKa = 3.86PADD558 pKa = 3.9GGTGGDD564 pKa = 3.59GNGWANDD571 pKa = 3.52PTTTNSRR578 pKa = 11.84QPDD581 pKa = 3.88PVSAFLPLVYY591 pKa = 10.18TPDD594 pKa = 4.04DD595 pKa = 4.21PQPTMLLLGLSDD607 pKa = 4.4PPAIAPEE614 pKa = 4.15TGQPEE619 pKa = 4.2RR620 pKa = 11.84RR621 pKa = 11.84FFDD624 pKa = 3.62LALITLGLISLTSGTFVRR642 pKa = 11.84FRR644 pKa = 11.84ARR646 pKa = 11.84RR647 pKa = 11.84VSS649 pKa = 3.11
MM1 pKa = 7.71RR2 pKa = 11.84RR3 pKa = 11.84VLWAIGLTLGLLSSLLFAGTAMAAGLGSVGTLSGAVSGVTDD44 pKa = 3.9TVSSVADD51 pKa = 3.56PVTATVSDD59 pKa = 3.85VAGAISDD66 pKa = 3.85AGSNGAGNNVNSGNTTSAGLIAVNDD91 pKa = 4.09LVDD94 pKa = 3.85VSALNGGNGGDD105 pKa = 3.3AGLNVSSPDD114 pKa = 3.34VTVGGLVAVNDD125 pKa = 4.03AVDD128 pKa = 3.46VSLLNGGNGDD138 pKa = 3.76GSGAGNNTNTGNTTTAGLIAVNDD161 pKa = 4.14LADD164 pKa = 3.47VSALNGANANGSHH177 pKa = 6.65AGSNAGNNTNGDD189 pKa = 3.67NTTSAGLIAVNDD201 pKa = 4.33GVDD204 pKa = 3.15ASLLNGANANGSHH217 pKa = 6.2SEE219 pKa = 3.99ANAGNNTNSGNTTSAGLVAVNDD241 pKa = 5.5GIDD244 pKa = 3.16ASLLNGANANGNHH257 pKa = 6.49AEE259 pKa = 4.1ANAGNNANSGNVTSGGLAGINDD281 pKa = 5.43GIDD284 pKa = 2.9ASFGNGANANGSLSGANAGNNVNTGNTTSSGVAGVNDD321 pKa = 5.59GIDD324 pKa = 3.2AGLLNGGNANGKK336 pKa = 8.24HH337 pKa = 5.77AEE339 pKa = 4.16VNAGNNVNTGNTTTGGLVGANDD361 pKa = 5.53LIDD364 pKa = 3.58ATLINGANANGDD376 pKa = 3.88HH377 pKa = 6.29AHH379 pKa = 7.01ANAGNNVNAGNTTSGGLIGINDD401 pKa = 4.46LLGATALNGANANGEE416 pKa = 4.17GARR419 pKa = 11.84ANAGNNINSGNITSGGLIAINDD441 pKa = 4.31PLDD444 pKa = 3.93LSLLNGANADD454 pKa = 3.59GSVGNHH460 pKa = 5.92GCTQNCGGDD469 pKa = 3.38NGCEE473 pKa = 3.9HH474 pKa = 7.79DD475 pKa = 4.09CGGDD479 pKa = 3.21HH480 pKa = 7.36GCGHH484 pKa = 7.57DD485 pKa = 3.51CGGDD489 pKa = 3.16HH490 pKa = 7.23GCTHH494 pKa = 6.61NCGGDD499 pKa = 3.64HH500 pKa = 6.46EE501 pKa = 5.28CSSNCGGNHH510 pKa = 7.04GGCTSNCSGNDD521 pKa = 3.27GHH523 pKa = 7.28CIANCEE529 pKa = 4.16GTSDD533 pKa = 3.49GGKK536 pKa = 10.09NNHH539 pKa = 6.54EE540 pKa = 4.47GNDD543 pKa = 4.13PIDD546 pKa = 4.16GGNHH550 pKa = 6.47NGGDD554 pKa = 3.84DD555 pKa = 3.86PADD558 pKa = 3.9GGTGGDD564 pKa = 3.59GNGWANDD571 pKa = 3.52PTTTNSRR578 pKa = 11.84QPDD581 pKa = 3.88PVSAFLPLVYY591 pKa = 10.18TPDD594 pKa = 4.04DD595 pKa = 4.21PQPTMLLLGLSDD607 pKa = 4.4PPAIAPEE614 pKa = 4.15TGQPEE619 pKa = 4.2RR620 pKa = 11.84RR621 pKa = 11.84FFDD624 pKa = 3.62LALITLGLISLTSGTFVRR642 pKa = 11.84FRR644 pKa = 11.84ARR646 pKa = 11.84RR647 pKa = 11.84VSS649 pKa = 3.11
Molecular weight: 62.13 kDa
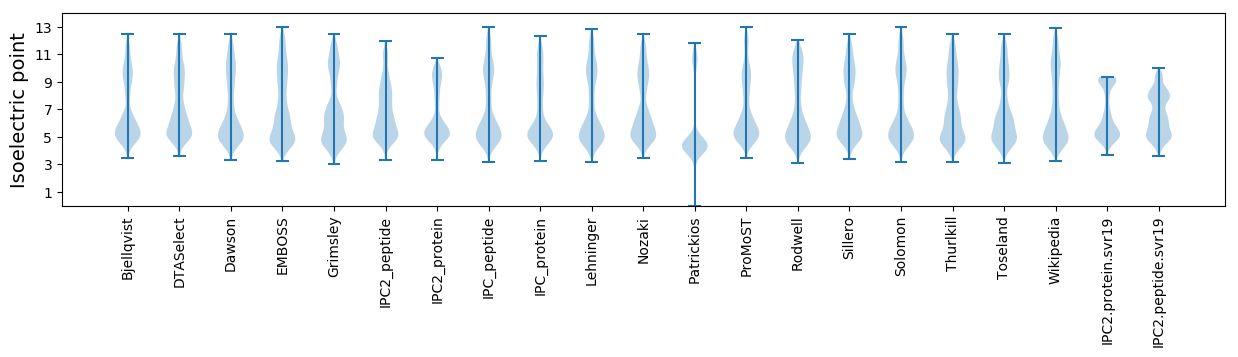
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I4ECS4|I4ECS4_9CHLR Formamidopyrimidine-DNA glycosylase OS=Nitrolancea hollandica Lb OX=1129897 GN=mutM PE=3 SV=1
MM1 pKa = 7.37LVCSGGVVVALLALSSSATFSGKK24 pKa = 9.97RR25 pKa = 11.84STATRR30 pKa = 11.84STEE33 pKa = 3.81DD34 pKa = 3.17TAGGTTHH41 pKa = 7.36ALSSPAHH48 pKa = 6.34PAGNPPRR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84DD58 pKa = 3.42LEE60 pKa = 4.23VTFRR64 pKa = 11.84RR65 pKa = 11.84LGLKK69 pKa = 9.56HH70 pKa = 6.34GRR72 pKa = 11.84IHH74 pKa = 7.38LPYY77 pKa = 10.29KK78 pKa = 10.32RR79 pKa = 11.84VLEE82 pKa = 4.23PAPATAMPPVLRR94 pKa = 11.84VSAQAPVPPIPMLGVSCGPRR114 pKa = 11.84MVMDD118 pKa = 4.75HH119 pKa = 7.14PPGALCYY126 pKa = 10.37
MM1 pKa = 7.37LVCSGGVVVALLALSSSATFSGKK24 pKa = 9.97RR25 pKa = 11.84STATRR30 pKa = 11.84STEE33 pKa = 3.81DD34 pKa = 3.17TAGGTTHH41 pKa = 7.36ALSSPAHH48 pKa = 6.34PAGNPPRR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84DD58 pKa = 3.42LEE60 pKa = 4.23VTFRR64 pKa = 11.84RR65 pKa = 11.84LGLKK69 pKa = 9.56HH70 pKa = 6.34GRR72 pKa = 11.84IHH74 pKa = 7.38LPYY77 pKa = 10.29KK78 pKa = 10.32RR79 pKa = 11.84VLEE82 pKa = 4.23PAPATAMPPVLRR94 pKa = 11.84VSAQAPVPPIPMLGVSCGPRR114 pKa = 11.84MVMDD118 pKa = 4.75HH119 pKa = 7.14PPGALCYY126 pKa = 10.37
Molecular weight: 13.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
962534 |
10 |
3045 |
242.5 |
26.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.803 ± 0.042 | 0.828 ± 0.014 |
5.434 ± 0.033 | 5.975 ± 0.045 |
3.343 ± 0.026 | 8.402 ± 0.042 |
2.225 ± 0.022 | 5.443 ± 0.034 |
2.356 ± 0.026 | 10.369 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.122 ± 0.02 | 2.512 ± 0.025 |
5.834 ± 0.032 | 3.584 ± 0.027 |
7.946 ± 0.05 | 5.561 ± 0.031 |
5.561 ± 0.037 | 7.686 ± 0.039 |
1.539 ± 0.019 | 2.476 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
