
Candida parapsilosis (strain CDC 317 / ATCC MYA-4646) (Yeast) (Monilia parapsilosis)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Debaryomycetaceae; Candida/Lodderomyces clade; Candida; Candida parapsilosis
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
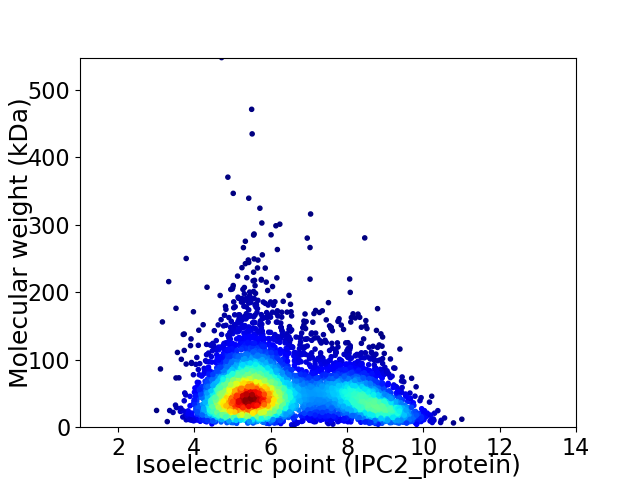
Virtual 2D-PAGE plot for 5777 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8B757|G8B757_CANPC Histone H2A OS=Candida parapsilosis (strain CDC 317 / ATCC MYA-4646) OX=578454 GN=CPAR2_103280 PE=3 SV=1
MM1 pKa = 7.76GDD3 pKa = 3.66HH4 pKa = 6.45VSEE7 pKa = 4.3IAWDD11 pKa = 3.88KK12 pKa = 11.22VDD14 pKa = 4.97SALTSNLLSKK24 pKa = 9.3ITNSLEE30 pKa = 3.79CSICSEE36 pKa = 4.01IMLAPMTTEE45 pKa = 4.74CGHH48 pKa = 5.47SFCYY52 pKa = 10.07EE53 pKa = 4.25CLHH56 pKa = 6.24QWFQNKK62 pKa = 8.68INCPTCRR69 pKa = 11.84HH70 pKa = 6.53EE71 pKa = 4.6IQTKK75 pKa = 9.13PALNMKK81 pKa = 10.29LNDD84 pKa = 3.48VSKK87 pKa = 11.09SLAEE91 pKa = 4.41LIIDD95 pKa = 4.27ARR97 pKa = 11.84LDD99 pKa = 3.63PNVDD103 pKa = 2.93SFRR106 pKa = 11.84ARR108 pKa = 11.84KK109 pKa = 8.93KK110 pKa = 10.65DD111 pKa = 3.36AMSKK115 pKa = 10.44YY116 pKa = 9.64EE117 pKa = 3.79SHH119 pKa = 6.67AKK121 pKa = 9.25KK122 pKa = 10.32KK123 pKa = 10.47RR124 pKa = 11.84VFGEE128 pKa = 4.0LFCGYY133 pKa = 10.23SLALIDD139 pKa = 5.68DD140 pKa = 3.97SDD142 pKa = 3.95GVPRR146 pKa = 11.84CGNCLWEE153 pKa = 4.08AHH155 pKa = 5.69GTVCLHH161 pKa = 6.31CGSRR165 pKa = 11.84FRR167 pKa = 11.84NTDD170 pKa = 4.3LIRR173 pKa = 11.84GGGHH177 pKa = 7.61DD178 pKa = 5.98DD179 pKa = 5.89DD180 pKa = 7.59DD181 pKa = 7.46DD182 pKa = 6.48DD183 pKa = 7.35DD184 pKa = 4.77EE185 pKa = 6.8DD186 pKa = 4.68EE187 pKa = 4.52EE188 pKa = 5.23EE189 pKa = 4.96FYY191 pKa = 11.28QDD193 pKa = 3.15VPPAEE198 pKa = 4.21DD199 pKa = 4.88HH200 pKa = 6.77YY201 pKa = 11.83DD202 pKa = 3.55SDD204 pKa = 5.1DD205 pKa = 3.97SFVDD209 pKa = 3.21SRR211 pKa = 11.84TAEE214 pKa = 3.95EE215 pKa = 4.22LLEE218 pKa = 4.72EE219 pKa = 4.12IHH221 pKa = 7.93DD222 pKa = 5.0NGPEE226 pKa = 4.31DD227 pKa = 5.24NNDD230 pKa = 3.6DD231 pKa = 3.92QPNATSSDD239 pKa = 3.43EE240 pKa = 5.04DD241 pKa = 4.84DD242 pKa = 5.26DD243 pKa = 6.74DD244 pKa = 4.53GAASFRR250 pKa = 11.84NWLSGSQPRR259 pKa = 11.84SHH261 pKa = 7.49IIYY264 pKa = 10.49VDD266 pKa = 3.57DD267 pKa = 3.88TDD269 pKa = 6.09SEE271 pKa = 4.56FEE273 pKa = 4.24YY274 pKa = 11.1NPRR277 pKa = 11.84IYY279 pKa = 10.65SDD281 pKa = 3.63SDD283 pKa = 3.26EE284 pKa = 5.75DD285 pKa = 4.1SPQQIHH291 pKa = 6.59FGGGPGGHH299 pKa = 6.58VIIDD303 pKa = 4.04LDD305 pKa = 4.02ADD307 pKa = 3.76EE308 pKa = 5.85DD309 pKa = 4.63GNDD312 pKa = 3.87SEE314 pKa = 6.37LIDD317 pKa = 4.09ALADD321 pKa = 3.91FQHH324 pKa = 6.41GVSEE328 pKa = 4.59DD329 pKa = 3.52EE330 pKa = 4.19ANEE333 pKa = 4.08DD334 pKa = 3.54FRR336 pKa = 11.84SLHH339 pKa = 6.4EE340 pKa = 4.14DD341 pKa = 3.54HH342 pKa = 7.24EE343 pKa = 4.56NDD345 pKa = 3.55YY346 pKa = 11.42YY347 pKa = 11.45DD348 pKa = 5.68RR349 pKa = 11.84SDD351 pKa = 3.68DD352 pKa = 4.25DD353 pKa = 4.09EE354 pKa = 6.85GGEE357 pKa = 4.19EE358 pKa = 4.93DD359 pKa = 3.75EE360 pKa = 5.6DD361 pKa = 4.2GHH363 pKa = 7.05GSHH366 pKa = 7.09SDD368 pKa = 3.61PDD370 pKa = 3.81PDD372 pKa = 4.19EE373 pKa = 5.76YY374 pKa = 11.61YY375 pKa = 10.99DD376 pKa = 4.34DD377 pKa = 5.28YY378 pKa = 11.46EE379 pKa = 6.59AEE381 pKa = 4.44DD382 pKa = 4.23HH383 pKa = 7.69DD384 pKa = 5.08IDD386 pKa = 5.1QDD388 pKa = 4.13DD389 pKa = 3.87VDD391 pKa = 4.29VDD393 pKa = 3.56VDD395 pKa = 3.61QNEE398 pKa = 4.22YY399 pKa = 10.99EE400 pKa = 5.31DD401 pKa = 5.54YY402 pKa = 11.34DD403 pKa = 5.21DD404 pKa = 5.64NYY406 pKa = 11.05NDD408 pKa = 4.26NDD410 pKa = 4.67DD411 pKa = 4.34YY412 pKa = 12.07DD413 pKa = 5.48DD414 pKa = 4.38GQEE417 pKa = 4.56TDD419 pKa = 4.1GNWW422 pKa = 2.86
MM1 pKa = 7.76GDD3 pKa = 3.66HH4 pKa = 6.45VSEE7 pKa = 4.3IAWDD11 pKa = 3.88KK12 pKa = 11.22VDD14 pKa = 4.97SALTSNLLSKK24 pKa = 9.3ITNSLEE30 pKa = 3.79CSICSEE36 pKa = 4.01IMLAPMTTEE45 pKa = 4.74CGHH48 pKa = 5.47SFCYY52 pKa = 10.07EE53 pKa = 4.25CLHH56 pKa = 6.24QWFQNKK62 pKa = 8.68INCPTCRR69 pKa = 11.84HH70 pKa = 6.53EE71 pKa = 4.6IQTKK75 pKa = 9.13PALNMKK81 pKa = 10.29LNDD84 pKa = 3.48VSKK87 pKa = 11.09SLAEE91 pKa = 4.41LIIDD95 pKa = 4.27ARR97 pKa = 11.84LDD99 pKa = 3.63PNVDD103 pKa = 2.93SFRR106 pKa = 11.84ARR108 pKa = 11.84KK109 pKa = 8.93KK110 pKa = 10.65DD111 pKa = 3.36AMSKK115 pKa = 10.44YY116 pKa = 9.64EE117 pKa = 3.79SHH119 pKa = 6.67AKK121 pKa = 9.25KK122 pKa = 10.32KK123 pKa = 10.47RR124 pKa = 11.84VFGEE128 pKa = 4.0LFCGYY133 pKa = 10.23SLALIDD139 pKa = 5.68DD140 pKa = 3.97SDD142 pKa = 3.95GVPRR146 pKa = 11.84CGNCLWEE153 pKa = 4.08AHH155 pKa = 5.69GTVCLHH161 pKa = 6.31CGSRR165 pKa = 11.84FRR167 pKa = 11.84NTDD170 pKa = 4.3LIRR173 pKa = 11.84GGGHH177 pKa = 7.61DD178 pKa = 5.98DD179 pKa = 5.89DD180 pKa = 7.59DD181 pKa = 7.46DD182 pKa = 6.48DD183 pKa = 7.35DD184 pKa = 4.77EE185 pKa = 6.8DD186 pKa = 4.68EE187 pKa = 4.52EE188 pKa = 5.23EE189 pKa = 4.96FYY191 pKa = 11.28QDD193 pKa = 3.15VPPAEE198 pKa = 4.21DD199 pKa = 4.88HH200 pKa = 6.77YY201 pKa = 11.83DD202 pKa = 3.55SDD204 pKa = 5.1DD205 pKa = 3.97SFVDD209 pKa = 3.21SRR211 pKa = 11.84TAEE214 pKa = 3.95EE215 pKa = 4.22LLEE218 pKa = 4.72EE219 pKa = 4.12IHH221 pKa = 7.93DD222 pKa = 5.0NGPEE226 pKa = 4.31DD227 pKa = 5.24NNDD230 pKa = 3.6DD231 pKa = 3.92QPNATSSDD239 pKa = 3.43EE240 pKa = 5.04DD241 pKa = 4.84DD242 pKa = 5.26DD243 pKa = 6.74DD244 pKa = 4.53GAASFRR250 pKa = 11.84NWLSGSQPRR259 pKa = 11.84SHH261 pKa = 7.49IIYY264 pKa = 10.49VDD266 pKa = 3.57DD267 pKa = 3.88TDD269 pKa = 6.09SEE271 pKa = 4.56FEE273 pKa = 4.24YY274 pKa = 11.1NPRR277 pKa = 11.84IYY279 pKa = 10.65SDD281 pKa = 3.63SDD283 pKa = 3.26EE284 pKa = 5.75DD285 pKa = 4.1SPQQIHH291 pKa = 6.59FGGGPGGHH299 pKa = 6.58VIIDD303 pKa = 4.04LDD305 pKa = 4.02ADD307 pKa = 3.76EE308 pKa = 5.85DD309 pKa = 4.63GNDD312 pKa = 3.87SEE314 pKa = 6.37LIDD317 pKa = 4.09ALADD321 pKa = 3.91FQHH324 pKa = 6.41GVSEE328 pKa = 4.59DD329 pKa = 3.52EE330 pKa = 4.19ANEE333 pKa = 4.08DD334 pKa = 3.54FRR336 pKa = 11.84SLHH339 pKa = 6.4EE340 pKa = 4.14DD341 pKa = 3.54HH342 pKa = 7.24EE343 pKa = 4.56NDD345 pKa = 3.55YY346 pKa = 11.42YY347 pKa = 11.45DD348 pKa = 5.68RR349 pKa = 11.84SDD351 pKa = 3.68DD352 pKa = 4.25DD353 pKa = 4.09EE354 pKa = 6.85GGEE357 pKa = 4.19EE358 pKa = 4.93DD359 pKa = 3.75EE360 pKa = 5.6DD361 pKa = 4.2GHH363 pKa = 7.05GSHH366 pKa = 7.09SDD368 pKa = 3.61PDD370 pKa = 3.81PDD372 pKa = 4.19EE373 pKa = 5.76YY374 pKa = 11.61YY375 pKa = 10.99DD376 pKa = 4.34DD377 pKa = 5.28YY378 pKa = 11.46EE379 pKa = 6.59AEE381 pKa = 4.44DD382 pKa = 4.23HH383 pKa = 7.69DD384 pKa = 5.08IDD386 pKa = 5.1QDD388 pKa = 4.13DD389 pKa = 3.87VDD391 pKa = 4.29VDD393 pKa = 3.56VDD395 pKa = 3.61QNEE398 pKa = 4.22YY399 pKa = 10.99EE400 pKa = 5.31DD401 pKa = 5.54YY402 pKa = 11.34DD403 pKa = 5.21DD404 pKa = 5.64NYY406 pKa = 11.05NDD408 pKa = 4.26NDD410 pKa = 4.67DD411 pKa = 4.34YY412 pKa = 12.07DD413 pKa = 5.48DD414 pKa = 4.38GQEE417 pKa = 4.56TDD419 pKa = 4.1GNWW422 pKa = 2.86
Molecular weight: 47.78 kDa
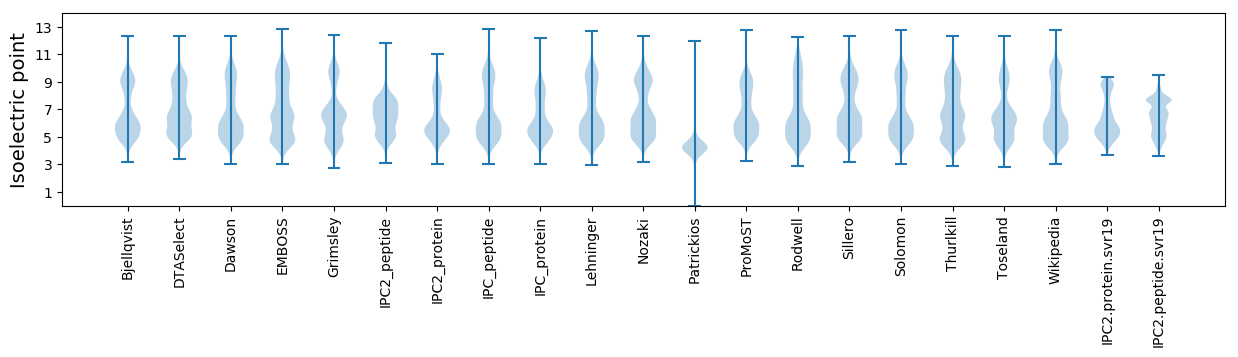
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8BAH0|G8BAH0_CANPC Uncharacterized protein OS=Candida parapsilosis (strain CDC 317 / ATCC MYA-4646) OX=578454 GN=CPAR2_805920 PE=4 SV=1
MM1 pKa = 7.26SLSRR5 pKa = 11.84PTTPIGDD12 pKa = 3.12RR13 pKa = 11.84RR14 pKa = 11.84QTNRR18 pKa = 11.84RR19 pKa = 11.84HH20 pKa = 5.15SRR22 pKa = 11.84RR23 pKa = 11.84SSGIPLPSQSPHH35 pKa = 6.91HH36 pKa = 6.44YY37 pKa = 10.12PVDD40 pKa = 4.0ADD42 pKa = 4.47NLPMDD47 pKa = 4.81CPEE50 pKa = 4.15IQNKK54 pKa = 9.31FKK56 pKa = 10.95DD57 pKa = 3.5IADD60 pKa = 3.92GLEE63 pKa = 4.13DD64 pKa = 4.6LDD66 pKa = 4.84VNVRR70 pKa = 11.84DD71 pKa = 4.56LNHH74 pKa = 6.03IHH76 pKa = 6.41NAVASQFNEE85 pKa = 3.97SFASFLYY92 pKa = 10.68GLSITMWCVDD102 pKa = 4.0LPKK105 pKa = 10.68CPSRR109 pKa = 11.84KK110 pKa = 9.2AWQKK114 pKa = 9.44LQMRR118 pKa = 11.84KK119 pKa = 9.24RR120 pKa = 11.84KK121 pKa = 9.51KK122 pKa = 8.69EE123 pKa = 3.73QIAKK127 pKa = 8.89VRR129 pKa = 11.84KK130 pKa = 8.67EE131 pKa = 3.59IEE133 pKa = 3.66GAEE136 pKa = 4.0RR137 pKa = 11.84LNAEE141 pKa = 4.6LKK143 pKa = 10.59EE144 pKa = 4.18KK145 pKa = 9.97VAKK148 pKa = 6.98EE149 pKa = 4.19TRR151 pKa = 11.84PIMYY155 pKa = 9.13SRR157 pKa = 11.84PMGTQLPKK165 pKa = 10.25RR166 pKa = 11.84ARR168 pKa = 11.84PNNEE172 pKa = 3.43NAKK175 pKa = 10.46PGINLNKK182 pKa = 9.51PSAPATRR189 pKa = 11.84VSRR192 pKa = 11.84IPQPVSGMLRR202 pKa = 11.84TQPGQTISRR211 pKa = 11.84NNAPNLDD218 pKa = 3.49QPPRR222 pKa = 11.84YY223 pKa = 9.16MRR225 pKa = 11.84GLFNNGVQSTPIGPNSRR242 pKa = 11.84VQKK245 pKa = 8.86PQRR248 pKa = 11.84AQHH251 pKa = 5.84SGRR254 pKa = 11.84RR255 pKa = 11.84PPFRR259 pKa = 5.79
MM1 pKa = 7.26SLSRR5 pKa = 11.84PTTPIGDD12 pKa = 3.12RR13 pKa = 11.84RR14 pKa = 11.84QTNRR18 pKa = 11.84RR19 pKa = 11.84HH20 pKa = 5.15SRR22 pKa = 11.84RR23 pKa = 11.84SSGIPLPSQSPHH35 pKa = 6.91HH36 pKa = 6.44YY37 pKa = 10.12PVDD40 pKa = 4.0ADD42 pKa = 4.47NLPMDD47 pKa = 4.81CPEE50 pKa = 4.15IQNKK54 pKa = 9.31FKK56 pKa = 10.95DD57 pKa = 3.5IADD60 pKa = 3.92GLEE63 pKa = 4.13DD64 pKa = 4.6LDD66 pKa = 4.84VNVRR70 pKa = 11.84DD71 pKa = 4.56LNHH74 pKa = 6.03IHH76 pKa = 6.41NAVASQFNEE85 pKa = 3.97SFASFLYY92 pKa = 10.68GLSITMWCVDD102 pKa = 4.0LPKK105 pKa = 10.68CPSRR109 pKa = 11.84KK110 pKa = 9.2AWQKK114 pKa = 9.44LQMRR118 pKa = 11.84KK119 pKa = 9.24RR120 pKa = 11.84KK121 pKa = 9.51KK122 pKa = 8.69EE123 pKa = 3.73QIAKK127 pKa = 8.89VRR129 pKa = 11.84KK130 pKa = 8.67EE131 pKa = 3.59IEE133 pKa = 3.66GAEE136 pKa = 4.0RR137 pKa = 11.84LNAEE141 pKa = 4.6LKK143 pKa = 10.59EE144 pKa = 4.18KK145 pKa = 9.97VAKK148 pKa = 6.98EE149 pKa = 4.19TRR151 pKa = 11.84PIMYY155 pKa = 9.13SRR157 pKa = 11.84PMGTQLPKK165 pKa = 10.25RR166 pKa = 11.84ARR168 pKa = 11.84PNNEE172 pKa = 3.43NAKK175 pKa = 10.46PGINLNKK182 pKa = 9.51PSAPATRR189 pKa = 11.84VSRR192 pKa = 11.84IPQPVSGMLRR202 pKa = 11.84TQPGQTISRR211 pKa = 11.84NNAPNLDD218 pKa = 3.49QPPRR222 pKa = 11.84YY223 pKa = 9.16MRR225 pKa = 11.84GLFNNGVQSTPIGPNSRR242 pKa = 11.84VQKK245 pKa = 8.86PQRR248 pKa = 11.84AQHH251 pKa = 5.84SGRR254 pKa = 11.84RR255 pKa = 11.84PPFRR259 pKa = 5.79
Molecular weight: 29.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2913513 |
33 |
4846 |
504.3 |
56.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.795 ± 0.027 | 1.085 ± 0.01 |
6.032 ± 0.024 | 6.477 ± 0.036 |
4.343 ± 0.022 | 5.486 ± 0.036 |
2.297 ± 0.013 | 6.315 ± 0.025 |
7.033 ± 0.031 | 9.121 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.861 ± 0.011 | 5.732 ± 0.024 |
4.537 ± 0.026 | 4.584 ± 0.03 |
4.144 ± 0.02 | 8.953 ± 0.044 |
5.873 ± 0.033 | 5.875 ± 0.021 |
1.002 ± 0.011 | 3.455 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
