
Botrytis elliptica
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Helotiales; Sclerotiniaceae; Botrytis
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
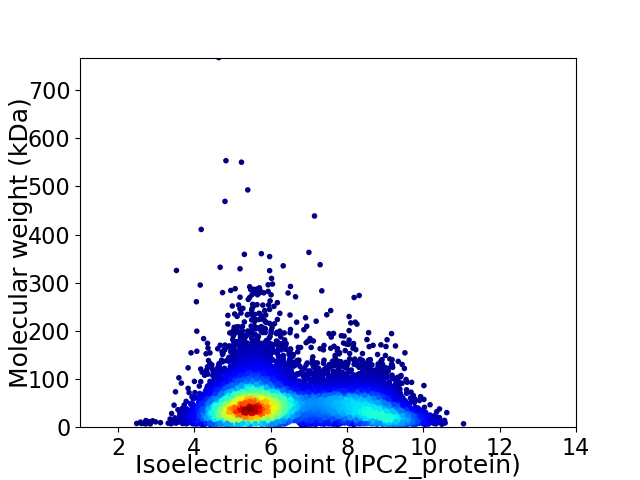
Virtual 2D-PAGE plot for 12555 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Z1JU93|A0A4Z1JU93_9HELO Uncharacterized protein OS=Botrytis elliptica OX=278938 GN=BELL_0452g00030 PE=4 SV=1
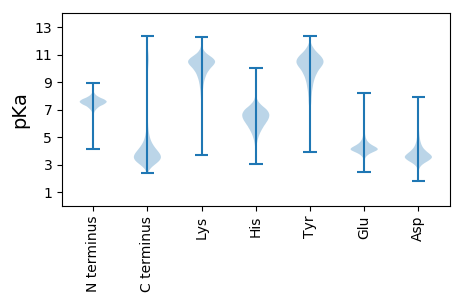
MM1 pKa = 7.74HH2 pKa = 7.34INILVLAASLCTVVLANYY20 pKa = 9.63PSDD23 pKa = 4.0DD24 pKa = 3.97NKK26 pKa = 11.26DD27 pKa = 3.36AVADD31 pKa = 3.88YY32 pKa = 10.36PFDD35 pKa = 4.81PNQAGVPDD43 pKa = 4.01VVLDD47 pKa = 4.12ASQAAVVSGIVSSLSSLSTALDD69 pKa = 3.49IPSSLASEE77 pKa = 4.94LSNIPTSASTLGYY90 pKa = 9.99SPTGVSQLDD99 pKa = 3.79EE100 pKa = 4.18QLSGTNKK107 pKa = 9.46PDD109 pKa = 2.98WYY111 pKa = 9.96TSLSPDD117 pKa = 2.95AKK119 pKa = 10.62SYY121 pKa = 11.09VDD123 pKa = 3.48NAVSILDD130 pKa = 3.54SVYY133 pKa = 10.83SLEE136 pKa = 5.24SILTTATAWPTTTVGVVGGTTATPLSTWEE165 pKa = 4.28STWASSTKK173 pKa = 10.02FDD175 pKa = 4.22DD176 pKa = 4.28WGSSWASSTGGASWDD191 pKa = 4.05SSWTTSTGADD201 pKa = 3.04SWDD204 pKa = 3.81SSWASSTGVNDD215 pKa = 3.86WEE217 pKa = 4.58STSPSGISSKK227 pKa = 10.77AGEE230 pKa = 4.47SVFGSSTGTKK240 pKa = 10.27DD241 pKa = 2.86STAAQTTGPIQVTTSNATPIMCNLRR266 pKa = 11.84LGFALAVAVGLICAMM281 pKa = 4.47
MM1 pKa = 7.74HH2 pKa = 7.34INILVLAASLCTVVLANYY20 pKa = 9.63PSDD23 pKa = 4.0DD24 pKa = 3.97NKK26 pKa = 11.26DD27 pKa = 3.36AVADD31 pKa = 3.88YY32 pKa = 10.36PFDD35 pKa = 4.81PNQAGVPDD43 pKa = 4.01VVLDD47 pKa = 4.12ASQAAVVSGIVSSLSSLSTALDD69 pKa = 3.49IPSSLASEE77 pKa = 4.94LSNIPTSASTLGYY90 pKa = 9.99SPTGVSQLDD99 pKa = 3.79EE100 pKa = 4.18QLSGTNKK107 pKa = 9.46PDD109 pKa = 2.98WYY111 pKa = 9.96TSLSPDD117 pKa = 2.95AKK119 pKa = 10.62SYY121 pKa = 11.09VDD123 pKa = 3.48NAVSILDD130 pKa = 3.54SVYY133 pKa = 10.83SLEE136 pKa = 5.24SILTTATAWPTTTVGVVGGTTATPLSTWEE165 pKa = 4.28STWASSTKK173 pKa = 10.02FDD175 pKa = 4.22DD176 pKa = 4.28WGSSWASSTGGASWDD191 pKa = 4.05SSWTTSTGADD201 pKa = 3.04SWDD204 pKa = 3.81SSWASSTGVNDD215 pKa = 3.86WEE217 pKa = 4.58STSPSGISSKK227 pKa = 10.77AGEE230 pKa = 4.47SVFGSSTGTKK240 pKa = 10.27DD241 pKa = 2.86STAAQTTGPIQVTTSNATPIMCNLRR266 pKa = 11.84LGFALAVAVGLICAMM281 pKa = 4.47
Molecular weight: 28.8 kDa
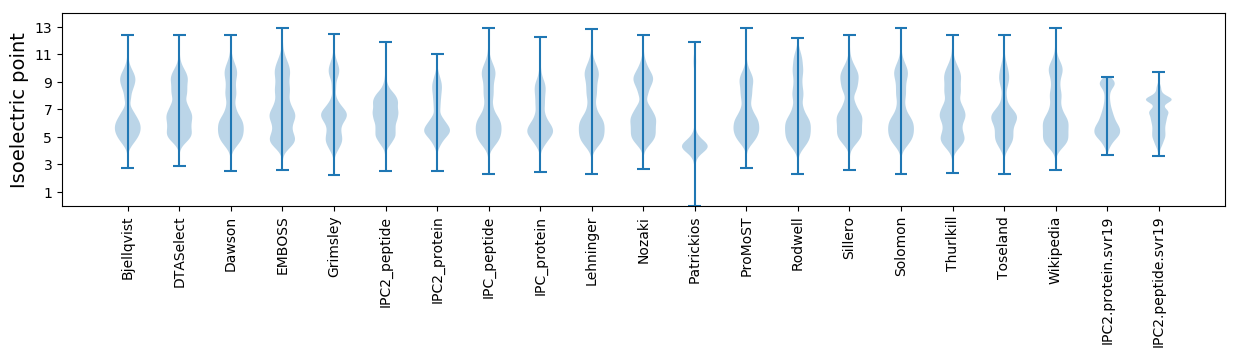
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Z1KGX3|A0A4Z1KGX3_9HELO Aminotran_5 domain-containing protein OS=Botrytis elliptica OX=278938 GN=BELL_0007g00130 PE=4 SV=1
MM1 pKa = 7.27SLSHH5 pKa = 5.73ATSWRR10 pKa = 11.84LRR12 pKa = 11.84PIEE15 pKa = 4.29SFNFHH20 pKa = 4.8TTKK23 pKa = 10.66RR24 pKa = 11.84PIPAPLKK31 pKa = 9.79PPPPKK36 pKa = 10.03PPPPKK41 pKa = 9.83PPPPKK46 pKa = 9.83PPPPKK51 pKa = 9.83PPPPKK56 pKa = 9.65PPTMTFSLTLTSLLHH71 pKa = 6.54LLPTLFGTIFILFGLNAMLRR91 pKa = 11.84PLHH94 pKa = 6.85ALTFYY99 pKa = 10.74PSLHH103 pKa = 6.58HH104 pKa = 6.64SLSPSLPQSPSNTILLEE121 pKa = 4.15SLLTIYY127 pKa = 10.32GARR130 pKa = 11.84DD131 pKa = 2.97IFMGPRR137 pKa = 11.84HH138 pKa = 5.81VRR140 pKa = 11.84RR141 pKa = 11.84ILPSQLQNTRR151 pKa = 11.84LDD153 pKa = 3.71RR154 pKa = 11.84GCGEE158 pKa = 4.2WCCVCGWVGVLEE170 pKa = 4.6GWRR173 pKa = 11.84GTGRR177 pKa = 11.84SLGVCARR184 pKa = 11.84TCGCGGFAGVGVLRR198 pKa = 11.84MII200 pKa = 5.03
MM1 pKa = 7.27SLSHH5 pKa = 5.73ATSWRR10 pKa = 11.84LRR12 pKa = 11.84PIEE15 pKa = 4.29SFNFHH20 pKa = 4.8TTKK23 pKa = 10.66RR24 pKa = 11.84PIPAPLKK31 pKa = 9.79PPPPKK36 pKa = 10.03PPPPKK41 pKa = 9.83PPPPKK46 pKa = 9.83PPPPKK51 pKa = 9.83PPPPKK56 pKa = 9.65PPTMTFSLTLTSLLHH71 pKa = 6.54LLPTLFGTIFILFGLNAMLRR91 pKa = 11.84PLHH94 pKa = 6.85ALTFYY99 pKa = 10.74PSLHH103 pKa = 6.58HH104 pKa = 6.64SLSPSLPQSPSNTILLEE121 pKa = 4.15SLLTIYY127 pKa = 10.32GARR130 pKa = 11.84DD131 pKa = 2.97IFMGPRR137 pKa = 11.84HH138 pKa = 5.81VRR140 pKa = 11.84RR141 pKa = 11.84ILPSQLQNTRR151 pKa = 11.84LDD153 pKa = 3.71RR154 pKa = 11.84GCGEE158 pKa = 4.2WCCVCGWVGVLEE170 pKa = 4.6GWRR173 pKa = 11.84GTGRR177 pKa = 11.84SLGVCARR184 pKa = 11.84TCGCGGFAGVGVLRR198 pKa = 11.84MII200 pKa = 5.03
Molecular weight: 21.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5947282 |
35 |
6915 |
473.7 |
52.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.666 ± 0.019 | 1.2 ± 0.009 |
5.586 ± 0.017 | 6.592 ± 0.025 |
3.731 ± 0.014 | 6.729 ± 0.019 |
2.284 ± 0.011 | 5.492 ± 0.015 |
5.472 ± 0.02 | 8.536 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.241 ± 0.009 | 4.262 ± 0.013 |
5.777 ± 0.022 | 3.881 ± 0.016 |
5.67 ± 0.019 | 8.785 ± 0.029 |
6.145 ± 0.019 | 5.73 ± 0.015 |
1.4 ± 0.009 | 2.821 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
