
Salipiger bermudensis (strain DSM 26914 / JCM 13377 / KCTC 12554 / HTCC2601) (Pelagibaca bermudensis)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Salipiger; Salipiger bermudensis
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
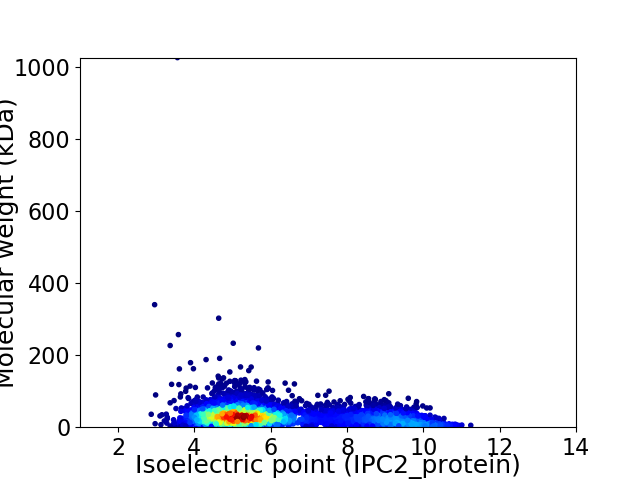
Virtual 2D-PAGE plot for 5420 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q0FL30|Q0FL30_SALBH (R)-citramalate synthase OS=Salipiger bermudensis (strain DSM 26914 / JCM 13377 / KCTC 12554 / HTCC2601) OX=314265 GN=R2601_23775 PE=3 SV=1
MM1 pKa = 7.93DD2 pKa = 5.81NDD4 pKa = 3.61VDD6 pKa = 3.8WFATQLDD13 pKa = 3.57ADD15 pKa = 3.55QYY17 pKa = 10.66YY18 pKa = 10.8RR19 pKa = 11.84IDD21 pKa = 4.32LVSASNGDD29 pKa = 3.63VLGLGDD35 pKa = 4.64PYY37 pKa = 11.34FLGLRR42 pKa = 11.84NSQGQIVYY50 pKa = 8.43GTGDD54 pKa = 3.57DD55 pKa = 5.2DD56 pKa = 5.19GGEE59 pKa = 4.76GYY61 pKa = 10.65NSRR64 pKa = 11.84SIFSVAQSGTYY75 pKa = 10.12YY76 pKa = 10.8LAASAFGTSTGAYY89 pKa = 8.09EE90 pKa = 4.75LNLTAYY96 pKa = 10.5DD97 pKa = 3.84FVDD100 pKa = 4.99DD101 pKa = 5.66FGADD105 pKa = 3.62PEE107 pKa = 4.6TSGNFSVGEE116 pKa = 4.25TVSGEE121 pKa = 3.87IQAGDD126 pKa = 3.61VYY128 pKa = 10.83QIQDD132 pKa = 3.99DD133 pKa = 3.82DD134 pKa = 3.72WFAIEE139 pKa = 4.04LTAGEE144 pKa = 4.72SYY146 pKa = 10.84IFDD149 pKa = 3.33VSASGNGTSDD159 pKa = 2.81SGGFPSIEE167 pKa = 3.63GLYY170 pKa = 10.42DD171 pKa = 3.42SEE173 pKa = 4.64GEE175 pKa = 4.88YY176 pKa = 10.59VSEE179 pKa = 4.26WNDD182 pKa = 3.08QVAYY186 pKa = 10.35RR187 pKa = 11.84GNKK190 pKa = 8.99VLFTPDD196 pKa = 2.88EE197 pKa = 4.46SGVYY201 pKa = 9.5HH202 pKa = 6.61VSVTNWDD209 pKa = 3.57YY210 pKa = 11.21FLEE213 pKa = 4.83GEE215 pKa = 4.88GGYY218 pKa = 9.93EE219 pKa = 4.04LQASIYY225 pKa = 10.24QRR227 pKa = 11.84IDD229 pKa = 3.1DD230 pKa = 4.47FAATVDD236 pKa = 3.39TTGILRR242 pKa = 11.84VGEE245 pKa = 4.1AATGEE250 pKa = 4.17IEE252 pKa = 5.52DD253 pKa = 3.85NAFLPRR259 pKa = 11.84ADD261 pKa = 4.22RR262 pKa = 11.84DD263 pKa = 3.37WFAVEE268 pKa = 4.29LEE270 pKa = 4.26EE271 pKa = 5.3GGVYY275 pKa = 9.63QFVMSPTDD283 pKa = 3.35ATGSLSYY290 pKa = 10.52SAVIRR295 pKa = 11.84GIYY298 pKa = 9.96DD299 pKa = 3.23SEE301 pKa = 6.04GDD303 pKa = 4.63LIPVMVRR310 pKa = 11.84NSYY313 pKa = 10.36YY314 pKa = 10.73LSNSLFFAPEE324 pKa = 3.55ANGTYY329 pKa = 9.6FVSASAEE336 pKa = 3.98WVDD339 pKa = 3.53VYY341 pKa = 11.24GAYY344 pKa = 9.91EE345 pKa = 4.01ISAEE349 pKa = 4.19RR350 pKa = 11.84IYY352 pKa = 11.34SFDD355 pKa = 3.44GYY357 pKa = 11.48VPDD360 pKa = 4.65AVASIQIHH368 pKa = 6.48AYY370 pKa = 8.8SPNYY374 pKa = 9.81VDD376 pKa = 5.22DD377 pKa = 4.8ASSTLVFGSDD387 pKa = 3.1RR388 pKa = 11.84DD389 pKa = 3.7NTIRR393 pKa = 11.84GISGHH398 pKa = 6.66DD399 pKa = 3.31VLLGAEE405 pKa = 4.68GDD407 pKa = 4.06DD408 pKa = 5.14LLIGDD413 pKa = 4.85DD414 pKa = 3.83TADD417 pKa = 4.0GQWLPGIEE425 pKa = 3.85GAVFRR430 pKa = 11.84LFEE433 pKa = 4.13FLLDD437 pKa = 3.78RR438 pKa = 11.84EE439 pKa = 4.15PSEE442 pKa = 5.98RR443 pKa = 11.84EE444 pKa = 3.64LQMGLEE450 pKa = 3.9NAYY453 pKa = 10.61YY454 pKa = 10.4SFYY457 pKa = 11.03GLSDD461 pKa = 3.33AAAFIIGSPEE471 pKa = 3.49FDD473 pKa = 3.58QRR475 pKa = 11.84WGDD478 pKa = 3.37LHH480 pKa = 6.87GADD483 pKa = 5.0LISFFYY489 pKa = 10.64EE490 pKa = 3.66RR491 pKa = 11.84LRR493 pKa = 11.84GNVVDD498 pKa = 4.69DD499 pKa = 3.99DD500 pKa = 4.49PYY502 pKa = 11.37RR503 pKa = 11.84EE504 pKa = 3.73IWLDD508 pKa = 3.43QLEE511 pKa = 4.38RR512 pKa = 11.84AYY514 pKa = 10.99SWGLLPRR521 pKa = 11.84YY522 pKa = 10.0AEE524 pKa = 4.31GEE526 pKa = 3.96IASAFVLSEE535 pKa = 4.45EE536 pKa = 4.37YY537 pKa = 10.29QSQSSGYY544 pKa = 9.44DD545 pKa = 2.97LTGRR549 pKa = 11.84LVEE552 pKa = 6.29FEE554 pKa = 4.39NQWGDD559 pKa = 3.69EE560 pKa = 4.33VFSSYY565 pKa = 9.75QAVLGRR571 pKa = 11.84RR572 pKa = 11.84PDD574 pKa = 3.23ARR576 pKa = 11.84GFEE579 pKa = 3.99NWLDD583 pKa = 4.1RR584 pKa = 11.84IEE586 pKa = 4.73NGLDD590 pKa = 3.32LSGLTRR596 pKa = 11.84MLLTSQEE603 pKa = 4.46FEE605 pKa = 4.02QSASGLSDD613 pKa = 3.27RR614 pKa = 11.84DD615 pKa = 3.55FVEE618 pKa = 5.5LVLTNLDD625 pKa = 3.67DD626 pKa = 3.8TTSEE630 pKa = 4.38VEE632 pKa = 3.94VDD634 pKa = 4.58FAAAQLDD641 pKa = 3.82AGLSRR646 pKa = 11.84DD647 pKa = 3.48EE648 pKa = 4.37FVAVQVLLNDD658 pKa = 4.01DD659 pKa = 3.83QGFDD663 pKa = 2.49AWMRR667 pKa = 11.84ARR669 pKa = 11.84GEE671 pKa = 4.13DD672 pKa = 3.66DD673 pKa = 3.6VLAGGAGNDD682 pKa = 4.16LLFGGPGSDD691 pKa = 2.47RR692 pKa = 11.84FVFAPDD698 pKa = 3.37EE699 pKa = 4.71GNDD702 pKa = 3.57TVADD706 pKa = 4.09LQSFDD711 pKa = 5.14IIDD714 pKa = 3.72LMAFGFADD722 pKa = 4.21LDD724 pKa = 3.76AALEE728 pKa = 4.28AFHH731 pKa = 6.82QDD733 pKa = 2.49GDD735 pKa = 4.14HH736 pKa = 6.99LVFEE740 pKa = 5.12AEE742 pKa = 3.82NTRR745 pKa = 11.84IRR747 pKa = 11.84FQSEE751 pKa = 3.44QLAALTDD758 pKa = 3.81DD759 pKa = 4.01MLQVV763 pKa = 3.39
MM1 pKa = 7.93DD2 pKa = 5.81NDD4 pKa = 3.61VDD6 pKa = 3.8WFATQLDD13 pKa = 3.57ADD15 pKa = 3.55QYY17 pKa = 10.66YY18 pKa = 10.8RR19 pKa = 11.84IDD21 pKa = 4.32LVSASNGDD29 pKa = 3.63VLGLGDD35 pKa = 4.64PYY37 pKa = 11.34FLGLRR42 pKa = 11.84NSQGQIVYY50 pKa = 8.43GTGDD54 pKa = 3.57DD55 pKa = 5.2DD56 pKa = 5.19GGEE59 pKa = 4.76GYY61 pKa = 10.65NSRR64 pKa = 11.84SIFSVAQSGTYY75 pKa = 10.12YY76 pKa = 10.8LAASAFGTSTGAYY89 pKa = 8.09EE90 pKa = 4.75LNLTAYY96 pKa = 10.5DD97 pKa = 3.84FVDD100 pKa = 4.99DD101 pKa = 5.66FGADD105 pKa = 3.62PEE107 pKa = 4.6TSGNFSVGEE116 pKa = 4.25TVSGEE121 pKa = 3.87IQAGDD126 pKa = 3.61VYY128 pKa = 10.83QIQDD132 pKa = 3.99DD133 pKa = 3.82DD134 pKa = 3.72WFAIEE139 pKa = 4.04LTAGEE144 pKa = 4.72SYY146 pKa = 10.84IFDD149 pKa = 3.33VSASGNGTSDD159 pKa = 2.81SGGFPSIEE167 pKa = 3.63GLYY170 pKa = 10.42DD171 pKa = 3.42SEE173 pKa = 4.64GEE175 pKa = 4.88YY176 pKa = 10.59VSEE179 pKa = 4.26WNDD182 pKa = 3.08QVAYY186 pKa = 10.35RR187 pKa = 11.84GNKK190 pKa = 8.99VLFTPDD196 pKa = 2.88EE197 pKa = 4.46SGVYY201 pKa = 9.5HH202 pKa = 6.61VSVTNWDD209 pKa = 3.57YY210 pKa = 11.21FLEE213 pKa = 4.83GEE215 pKa = 4.88GGYY218 pKa = 9.93EE219 pKa = 4.04LQASIYY225 pKa = 10.24QRR227 pKa = 11.84IDD229 pKa = 3.1DD230 pKa = 4.47FAATVDD236 pKa = 3.39TTGILRR242 pKa = 11.84VGEE245 pKa = 4.1AATGEE250 pKa = 4.17IEE252 pKa = 5.52DD253 pKa = 3.85NAFLPRR259 pKa = 11.84ADD261 pKa = 4.22RR262 pKa = 11.84DD263 pKa = 3.37WFAVEE268 pKa = 4.29LEE270 pKa = 4.26EE271 pKa = 5.3GGVYY275 pKa = 9.63QFVMSPTDD283 pKa = 3.35ATGSLSYY290 pKa = 10.52SAVIRR295 pKa = 11.84GIYY298 pKa = 9.96DD299 pKa = 3.23SEE301 pKa = 6.04GDD303 pKa = 4.63LIPVMVRR310 pKa = 11.84NSYY313 pKa = 10.36YY314 pKa = 10.73LSNSLFFAPEE324 pKa = 3.55ANGTYY329 pKa = 9.6FVSASAEE336 pKa = 3.98WVDD339 pKa = 3.53VYY341 pKa = 11.24GAYY344 pKa = 9.91EE345 pKa = 4.01ISAEE349 pKa = 4.19RR350 pKa = 11.84IYY352 pKa = 11.34SFDD355 pKa = 3.44GYY357 pKa = 11.48VPDD360 pKa = 4.65AVASIQIHH368 pKa = 6.48AYY370 pKa = 8.8SPNYY374 pKa = 9.81VDD376 pKa = 5.22DD377 pKa = 4.8ASSTLVFGSDD387 pKa = 3.1RR388 pKa = 11.84DD389 pKa = 3.7NTIRR393 pKa = 11.84GISGHH398 pKa = 6.66DD399 pKa = 3.31VLLGAEE405 pKa = 4.68GDD407 pKa = 4.06DD408 pKa = 5.14LLIGDD413 pKa = 4.85DD414 pKa = 3.83TADD417 pKa = 4.0GQWLPGIEE425 pKa = 3.85GAVFRR430 pKa = 11.84LFEE433 pKa = 4.13FLLDD437 pKa = 3.78RR438 pKa = 11.84EE439 pKa = 4.15PSEE442 pKa = 5.98RR443 pKa = 11.84EE444 pKa = 3.64LQMGLEE450 pKa = 3.9NAYY453 pKa = 10.61YY454 pKa = 10.4SFYY457 pKa = 11.03GLSDD461 pKa = 3.33AAAFIIGSPEE471 pKa = 3.49FDD473 pKa = 3.58QRR475 pKa = 11.84WGDD478 pKa = 3.37LHH480 pKa = 6.87GADD483 pKa = 5.0LISFFYY489 pKa = 10.64EE490 pKa = 3.66RR491 pKa = 11.84LRR493 pKa = 11.84GNVVDD498 pKa = 4.69DD499 pKa = 3.99DD500 pKa = 4.49PYY502 pKa = 11.37RR503 pKa = 11.84EE504 pKa = 3.73IWLDD508 pKa = 3.43QLEE511 pKa = 4.38RR512 pKa = 11.84AYY514 pKa = 10.99SWGLLPRR521 pKa = 11.84YY522 pKa = 10.0AEE524 pKa = 4.31GEE526 pKa = 3.96IASAFVLSEE535 pKa = 4.45EE536 pKa = 4.37YY537 pKa = 10.29QSQSSGYY544 pKa = 9.44DD545 pKa = 2.97LTGRR549 pKa = 11.84LVEE552 pKa = 6.29FEE554 pKa = 4.39NQWGDD559 pKa = 3.69EE560 pKa = 4.33VFSSYY565 pKa = 9.75QAVLGRR571 pKa = 11.84RR572 pKa = 11.84PDD574 pKa = 3.23ARR576 pKa = 11.84GFEE579 pKa = 3.99NWLDD583 pKa = 4.1RR584 pKa = 11.84IEE586 pKa = 4.73NGLDD590 pKa = 3.32LSGLTRR596 pKa = 11.84MLLTSQEE603 pKa = 4.46FEE605 pKa = 4.02QSASGLSDD613 pKa = 3.27RR614 pKa = 11.84DD615 pKa = 3.55FVEE618 pKa = 5.5LVLTNLDD625 pKa = 3.67DD626 pKa = 3.8TTSEE630 pKa = 4.38VEE632 pKa = 3.94VDD634 pKa = 4.58FAAAQLDD641 pKa = 3.82AGLSRR646 pKa = 11.84DD647 pKa = 3.48EE648 pKa = 4.37FVAVQVLLNDD658 pKa = 4.01DD659 pKa = 3.83QGFDD663 pKa = 2.49AWMRR667 pKa = 11.84ARR669 pKa = 11.84GEE671 pKa = 4.13DD672 pKa = 3.66DD673 pKa = 3.6VLAGGAGNDD682 pKa = 4.16LLFGGPGSDD691 pKa = 2.47RR692 pKa = 11.84FVFAPDD698 pKa = 3.37EE699 pKa = 4.71GNDD702 pKa = 3.57TVADD706 pKa = 4.09LQSFDD711 pKa = 5.14IIDD714 pKa = 3.72LMAFGFADD722 pKa = 4.21LDD724 pKa = 3.76AALEE728 pKa = 4.28AFHH731 pKa = 6.82QDD733 pKa = 2.49GDD735 pKa = 4.14HH736 pKa = 6.99LVFEE740 pKa = 5.12AEE742 pKa = 3.82NTRR745 pKa = 11.84IRR747 pKa = 11.84FQSEE751 pKa = 3.44QLAALTDD758 pKa = 3.81DD759 pKa = 4.01MLQVV763 pKa = 3.39
Molecular weight: 83.97 kDa
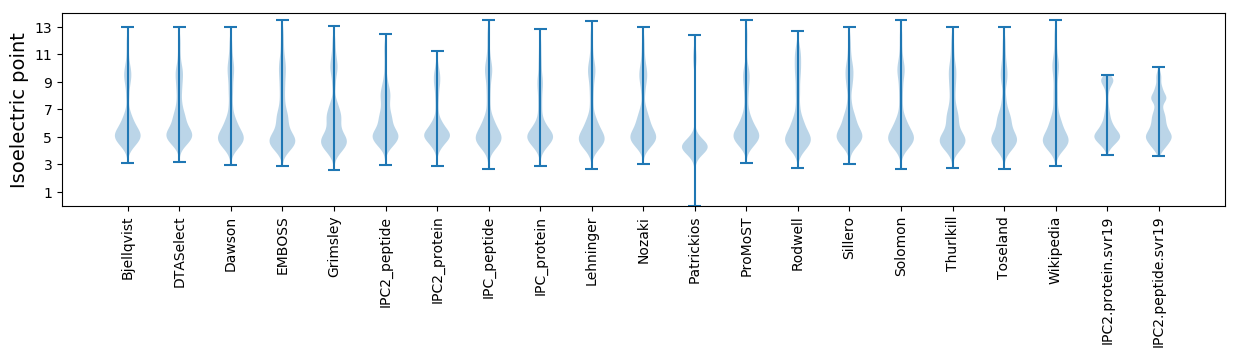
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q0FM68|Q0FM68_SALBH Pseudouridine synthase OS=Salipiger bermudensis (strain DSM 26914 / JCM 13377 / KCTC 12554 / HTCC2601) OX=314265 GN=R2601_17007 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84QILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84QILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1601589 |
16 |
10077 |
295.5 |
31.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.619 ± 0.045 | 0.901 ± 0.014 |
5.932 ± 0.048 | 6.361 ± 0.037 |
3.566 ± 0.019 | 8.906 ± 0.042 |
2.036 ± 0.022 | 4.873 ± 0.024 |
2.748 ± 0.027 | 10.27 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.779 ± 0.023 | 2.326 ± 0.027 |
5.271 ± 0.041 | 3.082 ± 0.018 |
6.947 ± 0.051 | 5.267 ± 0.037 |
5.393 ± 0.036 | 7.206 ± 0.031 |
1.383 ± 0.014 | 2.137 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
