
Gramella flava JLT2011
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Gramella; Gramella flava
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
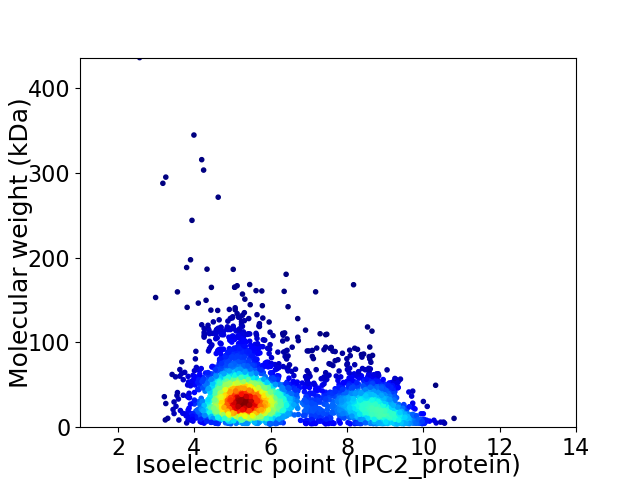
Virtual 2D-PAGE plot for 3628 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L7I558|A0A1L7I558_9FLAO Uncharacterized protein OS=Gramella flava JLT2011 OX=1229726 GN=GRFL_1521 PE=4 SV=1
MM1 pKa = 7.73KK2 pKa = 9.17EE3 pKa = 3.66LKK5 pKa = 9.98MIFSLLVVLLMFSGCDD21 pKa = 3.45GDD23 pKa = 5.58EE24 pKa = 5.44DD25 pKa = 4.75DD26 pKa = 5.6LQLEE30 pKa = 4.61SMVVPTNIQIDD41 pKa = 4.05TDD43 pKa = 3.34ISTDD47 pKa = 3.0GSGIVVFDD55 pKa = 3.7VSADD59 pKa = 3.35NAITYY64 pKa = 9.36KK65 pKa = 10.77FNFGDD70 pKa = 3.81EE71 pKa = 4.48TTGTSLDD78 pKa = 3.23GTYY81 pKa = 9.88TKK83 pKa = 10.25RR84 pKa = 11.84YY85 pKa = 8.61SRR87 pKa = 11.84NGLNTYY93 pKa = 9.4QVLVVAYY100 pKa = 8.96GKK102 pKa = 11.0GGISSSKK109 pKa = 9.87RR110 pKa = 11.84IEE112 pKa = 3.87ISVEE116 pKa = 3.82SDD118 pKa = 3.09FSDD121 pKa = 4.27PEE123 pKa = 4.19TKK125 pKa = 10.36EE126 pKa = 3.72FLTGGSSKK134 pKa = 8.61MWYY137 pKa = 9.12VAAFQPGHH145 pKa = 6.78LGVGPSSGDD154 pKa = 3.4GFDD157 pKa = 3.44SPIYY161 pKa = 10.08YY162 pKa = 10.08SAAPFEE168 pKa = 4.41KK169 pKa = 10.56AGAEE173 pKa = 3.84ASACFYY179 pKa = 10.93TDD181 pKa = 2.79VLTFSTTGEE190 pKa = 4.0NNFTYY195 pKa = 10.93NLNNNGATFFNVDD208 pKa = 3.17YY209 pKa = 10.42VGQFGGPTGSGDD221 pKa = 3.14QCIAYY226 pKa = 7.75DD227 pKa = 3.52TGGEE231 pKa = 4.18KK232 pKa = 9.6TVTLSPATSGIEE244 pKa = 4.12SSTGTVMDD252 pKa = 4.85FSDD255 pKa = 4.17GGFMSYY261 pKa = 11.04YY262 pKa = 9.75IGTSTYY268 pKa = 10.18EE269 pKa = 3.68ILDD272 pKa = 3.26IDD274 pKa = 4.43EE275 pKa = 4.3NSMYY279 pKa = 10.47VRR281 pKa = 11.84AIMGNNPALAWYY293 pKa = 9.91LKK295 pKa = 9.58FTTTPYY301 pKa = 10.68DD302 pKa = 3.49QQQEE306 pKa = 4.35SGNGGEE312 pKa = 4.47EE313 pKa = 3.75EE314 pKa = 5.37AEE316 pKa = 4.81FEE318 pKa = 4.5TQFNNLVWSDD328 pKa = 3.85EE329 pKa = 4.03FDD331 pKa = 4.12GDD333 pKa = 5.44SLDD336 pKa = 3.58TDD338 pKa = 3.14SWNFEE343 pKa = 4.33TGNGTNGWGNNEE355 pKa = 3.83LQYY358 pKa = 11.41YY359 pKa = 9.18LQDD362 pKa = 3.25NVTVQDD368 pKa = 3.52GSLFITAKK376 pKa = 10.29RR377 pKa = 11.84EE378 pKa = 4.17SEE380 pKa = 4.13SGFDD384 pKa = 3.58FTSGRR389 pKa = 11.84ITTQDD394 pKa = 3.06KK395 pKa = 11.46YY396 pKa = 9.91EE397 pKa = 3.96FTYY400 pKa = 10.82GRR402 pKa = 11.84VEE404 pKa = 3.95ARR406 pKa = 11.84IKK408 pKa = 10.75LPEE411 pKa = 4.45GVGTWPAFWMLGANFDD427 pKa = 3.94GVGWPEE433 pKa = 3.77TGEE436 pKa = 3.81IDD438 pKa = 3.19ILEE441 pKa = 4.41WVGKK445 pKa = 9.51EE446 pKa = 3.61PGRR449 pKa = 11.84IQSALHH455 pKa = 5.72FPGNSGGNAVVGPSTIEE472 pKa = 3.89NPSSEE477 pKa = 4.11FHH479 pKa = 6.74TYY481 pKa = 7.12TAEE484 pKa = 3.88WTEE487 pKa = 3.89DD488 pKa = 3.77AITFLLDD495 pKa = 3.11GEE497 pKa = 5.1VYY499 pKa = 8.92FTFDD503 pKa = 3.7NDD505 pKa = 3.22SDD507 pKa = 4.08KK508 pKa = 11.32PFNKK512 pKa = 10.21DD513 pKa = 2.79FFLILNVAMGGNLGGDD529 pKa = 3.35VDD531 pKa = 4.21PAFSEE536 pKa = 4.42SAMEE540 pKa = 3.65IDD542 pKa = 3.89YY543 pKa = 11.43VKK545 pKa = 10.7VYY547 pKa = 8.84QQ548 pKa = 4.21
MM1 pKa = 7.73KK2 pKa = 9.17EE3 pKa = 3.66LKK5 pKa = 9.98MIFSLLVVLLMFSGCDD21 pKa = 3.45GDD23 pKa = 5.58EE24 pKa = 5.44DD25 pKa = 4.75DD26 pKa = 5.6LQLEE30 pKa = 4.61SMVVPTNIQIDD41 pKa = 4.05TDD43 pKa = 3.34ISTDD47 pKa = 3.0GSGIVVFDD55 pKa = 3.7VSADD59 pKa = 3.35NAITYY64 pKa = 9.36KK65 pKa = 10.77FNFGDD70 pKa = 3.81EE71 pKa = 4.48TTGTSLDD78 pKa = 3.23GTYY81 pKa = 9.88TKK83 pKa = 10.25RR84 pKa = 11.84YY85 pKa = 8.61SRR87 pKa = 11.84NGLNTYY93 pKa = 9.4QVLVVAYY100 pKa = 8.96GKK102 pKa = 11.0GGISSSKK109 pKa = 9.87RR110 pKa = 11.84IEE112 pKa = 3.87ISVEE116 pKa = 3.82SDD118 pKa = 3.09FSDD121 pKa = 4.27PEE123 pKa = 4.19TKK125 pKa = 10.36EE126 pKa = 3.72FLTGGSSKK134 pKa = 8.61MWYY137 pKa = 9.12VAAFQPGHH145 pKa = 6.78LGVGPSSGDD154 pKa = 3.4GFDD157 pKa = 3.44SPIYY161 pKa = 10.08YY162 pKa = 10.08SAAPFEE168 pKa = 4.41KK169 pKa = 10.56AGAEE173 pKa = 3.84ASACFYY179 pKa = 10.93TDD181 pKa = 2.79VLTFSTTGEE190 pKa = 4.0NNFTYY195 pKa = 10.93NLNNNGATFFNVDD208 pKa = 3.17YY209 pKa = 10.42VGQFGGPTGSGDD221 pKa = 3.14QCIAYY226 pKa = 7.75DD227 pKa = 3.52TGGEE231 pKa = 4.18KK232 pKa = 9.6TVTLSPATSGIEE244 pKa = 4.12SSTGTVMDD252 pKa = 4.85FSDD255 pKa = 4.17GGFMSYY261 pKa = 11.04YY262 pKa = 9.75IGTSTYY268 pKa = 10.18EE269 pKa = 3.68ILDD272 pKa = 3.26IDD274 pKa = 4.43EE275 pKa = 4.3NSMYY279 pKa = 10.47VRR281 pKa = 11.84AIMGNNPALAWYY293 pKa = 9.91LKK295 pKa = 9.58FTTTPYY301 pKa = 10.68DD302 pKa = 3.49QQQEE306 pKa = 4.35SGNGGEE312 pKa = 4.47EE313 pKa = 3.75EE314 pKa = 5.37AEE316 pKa = 4.81FEE318 pKa = 4.5TQFNNLVWSDD328 pKa = 3.85EE329 pKa = 4.03FDD331 pKa = 4.12GDD333 pKa = 5.44SLDD336 pKa = 3.58TDD338 pKa = 3.14SWNFEE343 pKa = 4.33TGNGTNGWGNNEE355 pKa = 3.83LQYY358 pKa = 11.41YY359 pKa = 9.18LQDD362 pKa = 3.25NVTVQDD368 pKa = 3.52GSLFITAKK376 pKa = 10.29RR377 pKa = 11.84EE378 pKa = 4.17SEE380 pKa = 4.13SGFDD384 pKa = 3.58FTSGRR389 pKa = 11.84ITTQDD394 pKa = 3.06KK395 pKa = 11.46YY396 pKa = 9.91EE397 pKa = 3.96FTYY400 pKa = 10.82GRR402 pKa = 11.84VEE404 pKa = 3.95ARR406 pKa = 11.84IKK408 pKa = 10.75LPEE411 pKa = 4.45GVGTWPAFWMLGANFDD427 pKa = 3.94GVGWPEE433 pKa = 3.77TGEE436 pKa = 3.81IDD438 pKa = 3.19ILEE441 pKa = 4.41WVGKK445 pKa = 9.51EE446 pKa = 3.61PGRR449 pKa = 11.84IQSALHH455 pKa = 5.72FPGNSGGNAVVGPSTIEE472 pKa = 3.89NPSSEE477 pKa = 4.11FHH479 pKa = 6.74TYY481 pKa = 7.12TAEE484 pKa = 3.88WTEE487 pKa = 3.89DD488 pKa = 3.77AITFLLDD495 pKa = 3.11GEE497 pKa = 5.1VYY499 pKa = 8.92FTFDD503 pKa = 3.7NDD505 pKa = 3.22SDD507 pKa = 4.08KK508 pKa = 11.32PFNKK512 pKa = 10.21DD513 pKa = 2.79FFLILNVAMGGNLGGDD529 pKa = 3.35VDD531 pKa = 4.21PAFSEE536 pKa = 4.42SAMEE540 pKa = 3.65IDD542 pKa = 3.89YY543 pKa = 11.43VKK545 pKa = 10.7VYY547 pKa = 8.84QQ548 pKa = 4.21
Molecular weight: 59.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L7I2D2|A0A1L7I2D2_9FLAO Uncharacterized protein OS=Gramella flava JLT2011 OX=1229726 GN=GRFL_1017 PE=4 SV=1
MM1 pKa = 7.48SKK3 pKa = 9.86YY4 pKa = 10.32FRR6 pKa = 11.84YY7 pKa = 9.84FEE9 pKa = 3.78YY10 pKa = 10.54AYY12 pKa = 10.61LIFAAFFLFEE22 pKa = 5.43AVRR25 pKa = 11.84IWNSEE30 pKa = 3.55RR31 pKa = 11.84SRR33 pKa = 11.84AYY35 pKa = 9.41IFLFFVAIAVFMFFFKK51 pKa = 10.56RR52 pKa = 11.84RR53 pKa = 11.84FRR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84FEE59 pKa = 4.08EE60 pKa = 4.04RR61 pKa = 11.84NKK63 pKa = 11.0
MM1 pKa = 7.48SKK3 pKa = 9.86YY4 pKa = 10.32FRR6 pKa = 11.84YY7 pKa = 9.84FEE9 pKa = 3.78YY10 pKa = 10.54AYY12 pKa = 10.61LIFAAFFLFEE22 pKa = 5.43AVRR25 pKa = 11.84IWNSEE30 pKa = 3.55RR31 pKa = 11.84SRR33 pKa = 11.84AYY35 pKa = 9.41IFLFFVAIAVFMFFFKK51 pKa = 10.56RR52 pKa = 11.84RR53 pKa = 11.84FRR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84FEE59 pKa = 4.08EE60 pKa = 4.04RR61 pKa = 11.84NKK63 pKa = 11.0
Molecular weight: 8.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1223118 |
37 |
4291 |
337.1 |
38.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.605 ± 0.04 | 0.693 ± 0.019 |
5.715 ± 0.05 | 7.429 ± 0.039 |
5.211 ± 0.034 | 6.663 ± 0.043 |
1.81 ± 0.021 | 7.31 ± 0.038 |
7.08 ± 0.062 | 9.302 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.358 ± 0.026 | 5.595 ± 0.04 |
3.597 ± 0.024 | 3.788 ± 0.026 |
3.952 ± 0.032 | 6.519 ± 0.037 |
5.2 ± 0.058 | 6.041 ± 0.033 |
1.118 ± 0.016 | 4.015 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
