
Novosphingobium sp. 9U
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium; unclassified Novosphingobium
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
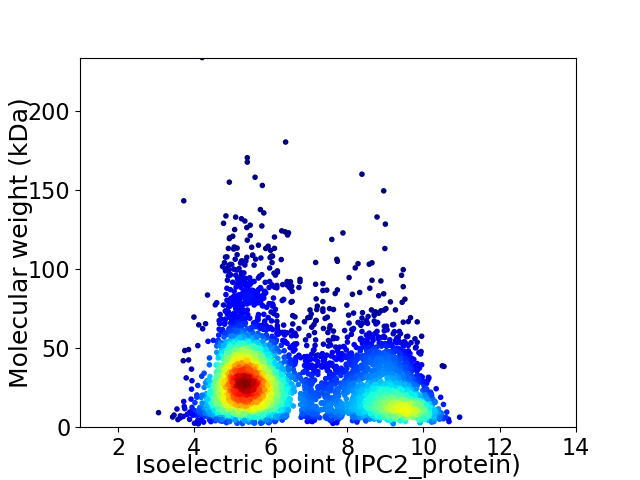
Virtual 2D-PAGE plot for 4782 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A653J8T0|A0A653J8T0_9SPHN Quinohemoprotein alcohol dehydrogenase ADH IIB OS=Novosphingobium sp. 9U OX=2653158 GN=qbdA PE=3 SV=1
MM1 pKa = 7.33TRR3 pKa = 11.84TFTSILALAALALPQAAPAATLLTNSDD30 pKa = 3.09GGYY33 pKa = 10.44GQIALPKK40 pKa = 10.02NDD42 pKa = 4.45DD43 pKa = 3.96SSSGAYY49 pKa = 9.39IMPFSIDD56 pKa = 3.56FFGSVYY62 pKa = 11.03DD63 pKa = 3.96EE64 pKa = 4.89LFVNNNGNVTFSTGLGSFTPFDD86 pKa = 4.35FGSLGQAMIAPFWADD101 pKa = 2.29VDD103 pKa = 4.4TRR105 pKa = 11.84CVSCGNVYY113 pKa = 10.18IGSFAPGQINVTWDD127 pKa = 3.11NVGYY131 pKa = 10.41YY132 pKa = 9.5SQHH135 pKa = 5.78SDD137 pKa = 3.19KK138 pKa = 11.43LNSFQLNLYY147 pKa = 9.97QSGNAGDD154 pKa = 3.99FDD156 pKa = 4.39IEE158 pKa = 4.66FRR160 pKa = 11.84YY161 pKa = 10.43ADD163 pKa = 4.29LNWTTGDD170 pKa = 3.2ASQGVNGLGGVPAAAGYY187 pKa = 8.75TNGAGATQLQPGSFLSPGALSFTDD211 pKa = 3.45SSNVGEE217 pKa = 4.51DD218 pKa = 3.45GRR220 pKa = 11.84WVYY223 pKa = 10.87NIRR226 pKa = 11.84NDD228 pKa = 4.99APPTDD233 pKa = 4.19YY234 pKa = 10.95GASPEE239 pKa = 4.19NPVLPTEE246 pKa = 4.04QTPGGGYY253 pKa = 10.41EE254 pKa = 4.08FTFEE258 pKa = 4.32IDD260 pKa = 3.1PTRR263 pKa = 11.84PTFIDD268 pKa = 3.82PPVATGYY275 pKa = 11.12DD276 pKa = 4.13FGAEE280 pKa = 3.83GGALFTSAVFTSLLNDD296 pKa = 3.17SDD298 pKa = 5.54GYY300 pKa = 10.57QLYY303 pKa = 10.89DD304 pKa = 3.31GANLLGQVAIGQTFTFASPLAAFQLRR330 pKa = 11.84GIDD333 pKa = 4.48SINMLQPGDD342 pKa = 3.74PLAFVSGFTFDD353 pKa = 2.9RR354 pKa = 11.84TGTVTVTQNPYY365 pKa = 9.12VQDD368 pKa = 3.31VAAAVPEE375 pKa = 4.23TATWAMMILGIGMVGGGLRR394 pKa = 11.84RR395 pKa = 11.84RR396 pKa = 11.84PPRR399 pKa = 11.84ARR401 pKa = 11.84FAA403 pKa = 4.07
MM1 pKa = 7.33TRR3 pKa = 11.84TFTSILALAALALPQAAPAATLLTNSDD30 pKa = 3.09GGYY33 pKa = 10.44GQIALPKK40 pKa = 10.02NDD42 pKa = 4.45DD43 pKa = 3.96SSSGAYY49 pKa = 9.39IMPFSIDD56 pKa = 3.56FFGSVYY62 pKa = 11.03DD63 pKa = 3.96EE64 pKa = 4.89LFVNNNGNVTFSTGLGSFTPFDD86 pKa = 4.35FGSLGQAMIAPFWADD101 pKa = 2.29VDD103 pKa = 4.4TRR105 pKa = 11.84CVSCGNVYY113 pKa = 10.18IGSFAPGQINVTWDD127 pKa = 3.11NVGYY131 pKa = 10.41YY132 pKa = 9.5SQHH135 pKa = 5.78SDD137 pKa = 3.19KK138 pKa = 11.43LNSFQLNLYY147 pKa = 9.97QSGNAGDD154 pKa = 3.99FDD156 pKa = 4.39IEE158 pKa = 4.66FRR160 pKa = 11.84YY161 pKa = 10.43ADD163 pKa = 4.29LNWTTGDD170 pKa = 3.2ASQGVNGLGGVPAAAGYY187 pKa = 8.75TNGAGATQLQPGSFLSPGALSFTDD211 pKa = 3.45SSNVGEE217 pKa = 4.51DD218 pKa = 3.45GRR220 pKa = 11.84WVYY223 pKa = 10.87NIRR226 pKa = 11.84NDD228 pKa = 4.99APPTDD233 pKa = 4.19YY234 pKa = 10.95GASPEE239 pKa = 4.19NPVLPTEE246 pKa = 4.04QTPGGGYY253 pKa = 10.41EE254 pKa = 4.08FTFEE258 pKa = 4.32IDD260 pKa = 3.1PTRR263 pKa = 11.84PTFIDD268 pKa = 3.82PPVATGYY275 pKa = 11.12DD276 pKa = 4.13FGAEE280 pKa = 3.83GGALFTSAVFTSLLNDD296 pKa = 3.17SDD298 pKa = 5.54GYY300 pKa = 10.57QLYY303 pKa = 10.89DD304 pKa = 3.31GANLLGQVAIGQTFTFASPLAAFQLRR330 pKa = 11.84GIDD333 pKa = 4.48SINMLQPGDD342 pKa = 3.74PLAFVSGFTFDD353 pKa = 2.9RR354 pKa = 11.84TGTVTVTQNPYY365 pKa = 9.12VQDD368 pKa = 3.31VAAAVPEE375 pKa = 4.23TATWAMMILGIGMVGGGLRR394 pKa = 11.84RR395 pKa = 11.84RR396 pKa = 11.84PPRR399 pKa = 11.84ARR401 pKa = 11.84FAA403 pKa = 4.07
Molecular weight: 42.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A653JB82|A0A653JB82_9SPHN Outer membrane-specific lipoprotein transporter subunit ATP-binding component of ABC superfamily OS=Novosphingobium sp. 9U OX=2653158 GN=lolD PE=4 SV=1
MM1 pKa = 7.56PRR3 pKa = 11.84PRR5 pKa = 11.84EE6 pKa = 3.7PASPFRR12 pKa = 11.84YY13 pKa = 9.43FISSPEE19 pKa = 4.07VIRR22 pKa = 11.84LVVLMYY28 pKa = 10.91VRR30 pKa = 11.84FPLSLRR36 pKa = 11.84NVEE39 pKa = 4.48DD40 pKa = 3.88LPFEE44 pKa = 4.39RR45 pKa = 11.84GIDD48 pKa = 3.48ICHH51 pKa = 5.07EE52 pKa = 4.15TARR55 pKa = 11.84LRR57 pKa = 11.84WNRR60 pKa = 11.84FGPMFAGEE68 pKa = 4.19VQRR71 pKa = 11.84KK72 pKa = 7.8RR73 pKa = 11.84VCRR76 pKa = 11.84MRR78 pKa = 11.84GLRR81 pKa = 11.84HH82 pKa = 5.56WRR84 pKa = 11.84WHH86 pKa = 5.88LDD88 pKa = 3.16QRR90 pKa = 11.84YY91 pKa = 9.54AKK93 pKa = 10.62LNDD96 pKa = 3.04EE97 pKa = 4.57MVYY100 pKa = 10.17LWRR103 pKa = 11.84AVDD106 pKa = 3.72HH107 pKa = 6.42EE108 pKa = 4.84GEE110 pKa = 4.26VLEE113 pKa = 4.63SYY115 pKa = 10.31IIKK118 pKa = 9.2TRR120 pKa = 11.84DD121 pKa = 2.87KK122 pKa = 10.87AAALSFMKK130 pKa = 10.44KK131 pKa = 9.41ALKK134 pKa = 10.28RR135 pKa = 11.84HH136 pKa = 6.1GSPEE140 pKa = 4.21TITTDD145 pKa = 3.93GLRR148 pKa = 11.84SCCAAMNEE156 pKa = 4.59LGNCEE161 pKa = 3.94KK162 pKa = 10.96QEE164 pKa = 3.91VGRR167 pKa = 11.84RR168 pKa = 11.84ANNRR172 pKa = 11.84VEE174 pKa = 4.38NSHH177 pKa = 6.79LPFRR181 pKa = 11.84RR182 pKa = 11.84RR183 pKa = 11.84KK184 pKa = 9.39SAMLRR189 pKa = 11.84FRR191 pKa = 11.84RR192 pKa = 11.84VKK194 pKa = 9.6TLQKK198 pKa = 10.08FASVHH203 pKa = 5.88ANIHH207 pKa = 5.04NHH209 pKa = 5.43FQPGTSPSQPRR220 pKa = 11.84DD221 pKa = 3.48LQGTPLSCLGG231 pKa = 3.65
MM1 pKa = 7.56PRR3 pKa = 11.84PRR5 pKa = 11.84EE6 pKa = 3.7PASPFRR12 pKa = 11.84YY13 pKa = 9.43FISSPEE19 pKa = 4.07VIRR22 pKa = 11.84LVVLMYY28 pKa = 10.91VRR30 pKa = 11.84FPLSLRR36 pKa = 11.84NVEE39 pKa = 4.48DD40 pKa = 3.88LPFEE44 pKa = 4.39RR45 pKa = 11.84GIDD48 pKa = 3.48ICHH51 pKa = 5.07EE52 pKa = 4.15TARR55 pKa = 11.84LRR57 pKa = 11.84WNRR60 pKa = 11.84FGPMFAGEE68 pKa = 4.19VQRR71 pKa = 11.84KK72 pKa = 7.8RR73 pKa = 11.84VCRR76 pKa = 11.84MRR78 pKa = 11.84GLRR81 pKa = 11.84HH82 pKa = 5.56WRR84 pKa = 11.84WHH86 pKa = 5.88LDD88 pKa = 3.16QRR90 pKa = 11.84YY91 pKa = 9.54AKK93 pKa = 10.62LNDD96 pKa = 3.04EE97 pKa = 4.57MVYY100 pKa = 10.17LWRR103 pKa = 11.84AVDD106 pKa = 3.72HH107 pKa = 6.42EE108 pKa = 4.84GEE110 pKa = 4.26VLEE113 pKa = 4.63SYY115 pKa = 10.31IIKK118 pKa = 9.2TRR120 pKa = 11.84DD121 pKa = 2.87KK122 pKa = 10.87AAALSFMKK130 pKa = 10.44KK131 pKa = 9.41ALKK134 pKa = 10.28RR135 pKa = 11.84HH136 pKa = 6.1GSPEE140 pKa = 4.21TITTDD145 pKa = 3.93GLRR148 pKa = 11.84SCCAAMNEE156 pKa = 4.59LGNCEE161 pKa = 3.94KK162 pKa = 10.96QEE164 pKa = 3.91VGRR167 pKa = 11.84RR168 pKa = 11.84ANNRR172 pKa = 11.84VEE174 pKa = 4.38NSHH177 pKa = 6.79LPFRR181 pKa = 11.84RR182 pKa = 11.84RR183 pKa = 11.84KK184 pKa = 9.39SAMLRR189 pKa = 11.84FRR191 pKa = 11.84RR192 pKa = 11.84VKK194 pKa = 9.6TLQKK198 pKa = 10.08FASVHH203 pKa = 5.88ANIHH207 pKa = 5.04NHH209 pKa = 5.43FQPGTSPSQPRR220 pKa = 11.84DD221 pKa = 3.48LQGTPLSCLGG231 pKa = 3.65
Molecular weight: 27.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1383869 |
21 |
2345 |
289.4 |
31.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.052 ± 0.058 | 0.92 ± 0.013 |
5.702 ± 0.025 | 5.531 ± 0.034 |
3.463 ± 0.022 | 8.793 ± 0.041 |
2.086 ± 0.022 | 4.661 ± 0.026 |
2.994 ± 0.028 | 9.953 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.396 ± 0.02 | 2.522 ± 0.023 |
5.345 ± 0.026 | 3.353 ± 0.02 |
7.369 ± 0.041 | 5.542 ± 0.027 |
5.324 ± 0.031 | 7.296 ± 0.03 |
1.45 ± 0.015 | 2.247 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
