
Albugo candida
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Albuginales; Albuginaceae; Albugo
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
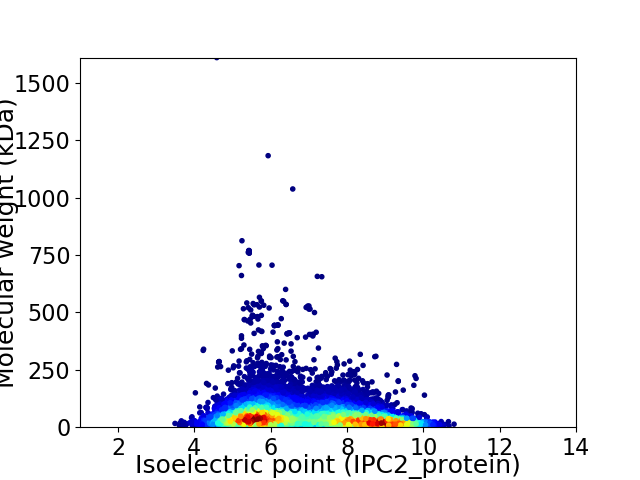
Virtual 2D-PAGE plot for 13200 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
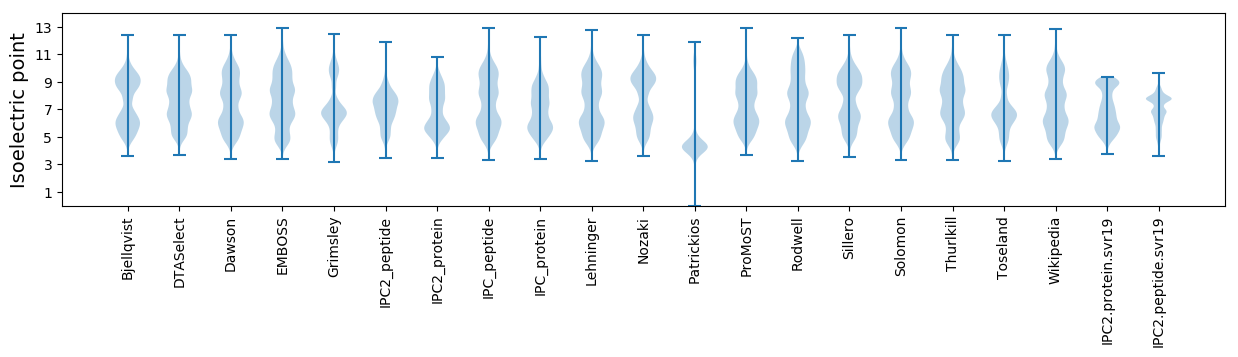
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A024GRH0|A0A024GRH0_9STRA Uncharacterized protein OS=Albugo candida OX=65357 GN=BN9_108320 PE=4 SV=1
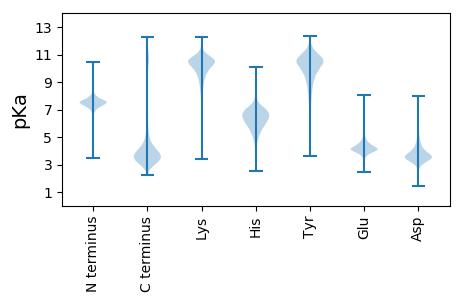
MM1 pKa = 7.82RR2 pKa = 11.84LLASLQMISVALTVIGVTTTLQEE25 pKa = 4.3VQSSSSSLRR34 pKa = 11.84SLQSSDD40 pKa = 2.55HH41 pKa = 6.03SEE43 pKa = 3.94RR44 pKa = 11.84NNRR47 pKa = 11.84PTISMVVISEE57 pKa = 4.09YY58 pKa = 10.9FPIPPTEE65 pKa = 4.27EE66 pKa = 4.18ADD68 pKa = 3.47QSQNEE73 pKa = 4.35PKK75 pKa = 10.64VVIPDD80 pKa = 3.26DD81 pKa = 3.99TFNEE85 pKa = 4.55KK86 pKa = 10.86YY87 pKa = 10.77GIFDD91 pKa = 3.77GGDD94 pKa = 3.35DD95 pKa = 3.78EE96 pKa = 5.63EE97 pKa = 4.88EE98 pKa = 4.1EE99 pKa = 4.43EE100 pKa = 4.36EE101 pKa = 4.47EE102 pKa = 4.36KK103 pKa = 11.22VHH105 pKa = 6.33TGYY108 pKa = 10.78EE109 pKa = 4.26LEE111 pKa = 4.47CEE113 pKa = 4.2GLEE116 pKa = 4.08KK117 pKa = 10.9DD118 pKa = 3.51SAEE121 pKa = 4.19SVNEE125 pKa = 3.62ISSDD129 pKa = 3.82YY130 pKa = 11.44LLDD133 pKa = 4.14CQDD136 pKa = 4.29DD137 pKa = 4.12EE138 pKa = 6.15KK139 pKa = 11.82VDD141 pKa = 4.58LDD143 pKa = 4.09TKK145 pKa = 10.87KK146 pKa = 8.7HH147 pKa = 4.97TKK149 pKa = 7.31EE150 pKa = 3.62TGYY153 pKa = 10.83GQTFFDD159 pKa = 5.58CDD161 pKa = 3.91DD162 pKa = 4.15EE163 pKa = 6.76GDD165 pKa = 3.92NVDD168 pKa = 5.03EE169 pKa = 4.31VVQDD173 pKa = 3.58KK174 pKa = 11.21EE175 pKa = 4.38EE176 pKa = 4.06NTEE179 pKa = 3.73EE180 pKa = 3.91MRR182 pKa = 11.84YY183 pKa = 9.41EE184 pKa = 4.07LQPLLDD190 pKa = 5.34CDD192 pKa = 6.17DD193 pKa = 4.15EE194 pKa = 7.03DD195 pKa = 4.61EE196 pKa = 6.04DD197 pKa = 5.3INEE200 pKa = 3.84VDD202 pKa = 3.51QDD204 pKa = 3.73KK205 pKa = 11.5EE206 pKa = 4.3EE207 pKa = 4.14NTEE210 pKa = 3.79ATRR213 pKa = 11.84YY214 pKa = 9.21NPQPILICDD223 pKa = 3.55EE224 pKa = 5.57DD225 pKa = 4.2EE226 pKa = 6.64DD227 pKa = 4.92EE228 pKa = 6.7DD229 pKa = 4.78IDD231 pKa = 4.09HH232 pKa = 7.24KK233 pKa = 11.14SNSMVLKK240 pKa = 10.59EE241 pKa = 4.04EE242 pKa = 4.17AKK244 pKa = 10.55SLQEE248 pKa = 4.46SKK250 pKa = 10.88RR251 pKa = 11.84EE252 pKa = 4.06EE253 pKa = 3.9NWSEE257 pKa = 3.45ILQYY261 pKa = 11.59DD262 pKa = 4.03SMQAYY267 pKa = 10.12LDD269 pKa = 3.79KK270 pKa = 10.79EE271 pKa = 4.33AEE273 pKa = 4.02KK274 pKa = 10.9SRR276 pKa = 11.84EE277 pKa = 3.91SEE279 pKa = 3.29FDD281 pKa = 3.71YY282 pKa = 11.55NFGRR286 pKa = 11.84SNDD289 pKa = 3.48NEE291 pKa = 3.83FRR293 pKa = 11.84SCKK296 pKa = 9.93SLCAYY301 pKa = 7.86DD302 pKa = 3.83TTGPACGSDD311 pKa = 2.86GQTYY315 pKa = 10.86SNEE318 pKa = 3.61CMMRR322 pKa = 11.84CLEE325 pKa = 4.27PDD327 pKa = 2.52IKK329 pKa = 10.78YY330 pKa = 10.17YY331 pKa = 10.83HH332 pKa = 7.06EE333 pKa = 5.57GYY335 pKa = 10.43CLQQEE340 pKa = 4.46DD341 pKa = 4.47SSCDD345 pKa = 3.2VDD347 pKa = 4.87CSHH350 pKa = 6.65THH352 pKa = 7.18DD353 pKa = 4.76LVCGIDD359 pKa = 3.2GQTYY363 pKa = 10.5INYY366 pKa = 9.58CLYY369 pKa = 11.02AVTYY373 pKa = 9.84CDD375 pKa = 4.6KK376 pKa = 11.2NLATLPFLFGACEE389 pKa = 4.06SDD391 pKa = 3.56KK392 pKa = 10.71VTAFF396 pKa = 3.46
MM1 pKa = 7.82RR2 pKa = 11.84LLASLQMISVALTVIGVTTTLQEE25 pKa = 4.3VQSSSSSLRR34 pKa = 11.84SLQSSDD40 pKa = 2.55HH41 pKa = 6.03SEE43 pKa = 3.94RR44 pKa = 11.84NNRR47 pKa = 11.84PTISMVVISEE57 pKa = 4.09YY58 pKa = 10.9FPIPPTEE65 pKa = 4.27EE66 pKa = 4.18ADD68 pKa = 3.47QSQNEE73 pKa = 4.35PKK75 pKa = 10.64VVIPDD80 pKa = 3.26DD81 pKa = 3.99TFNEE85 pKa = 4.55KK86 pKa = 10.86YY87 pKa = 10.77GIFDD91 pKa = 3.77GGDD94 pKa = 3.35DD95 pKa = 3.78EE96 pKa = 5.63EE97 pKa = 4.88EE98 pKa = 4.1EE99 pKa = 4.43EE100 pKa = 4.36EE101 pKa = 4.47EE102 pKa = 4.36KK103 pKa = 11.22VHH105 pKa = 6.33TGYY108 pKa = 10.78EE109 pKa = 4.26LEE111 pKa = 4.47CEE113 pKa = 4.2GLEE116 pKa = 4.08KK117 pKa = 10.9DD118 pKa = 3.51SAEE121 pKa = 4.19SVNEE125 pKa = 3.62ISSDD129 pKa = 3.82YY130 pKa = 11.44LLDD133 pKa = 4.14CQDD136 pKa = 4.29DD137 pKa = 4.12EE138 pKa = 6.15KK139 pKa = 11.82VDD141 pKa = 4.58LDD143 pKa = 4.09TKK145 pKa = 10.87KK146 pKa = 8.7HH147 pKa = 4.97TKK149 pKa = 7.31EE150 pKa = 3.62TGYY153 pKa = 10.83GQTFFDD159 pKa = 5.58CDD161 pKa = 3.91DD162 pKa = 4.15EE163 pKa = 6.76GDD165 pKa = 3.92NVDD168 pKa = 5.03EE169 pKa = 4.31VVQDD173 pKa = 3.58KK174 pKa = 11.21EE175 pKa = 4.38EE176 pKa = 4.06NTEE179 pKa = 3.73EE180 pKa = 3.91MRR182 pKa = 11.84YY183 pKa = 9.41EE184 pKa = 4.07LQPLLDD190 pKa = 5.34CDD192 pKa = 6.17DD193 pKa = 4.15EE194 pKa = 7.03DD195 pKa = 4.61EE196 pKa = 6.04DD197 pKa = 5.3INEE200 pKa = 3.84VDD202 pKa = 3.51QDD204 pKa = 3.73KK205 pKa = 11.5EE206 pKa = 4.3EE207 pKa = 4.14NTEE210 pKa = 3.79ATRR213 pKa = 11.84YY214 pKa = 9.21NPQPILICDD223 pKa = 3.55EE224 pKa = 5.57DD225 pKa = 4.2EE226 pKa = 6.64DD227 pKa = 4.92EE228 pKa = 6.7DD229 pKa = 4.78IDD231 pKa = 4.09HH232 pKa = 7.24KK233 pKa = 11.14SNSMVLKK240 pKa = 10.59EE241 pKa = 4.04EE242 pKa = 4.17AKK244 pKa = 10.55SLQEE248 pKa = 4.46SKK250 pKa = 10.88RR251 pKa = 11.84EE252 pKa = 4.06EE253 pKa = 3.9NWSEE257 pKa = 3.45ILQYY261 pKa = 11.59DD262 pKa = 4.03SMQAYY267 pKa = 10.12LDD269 pKa = 3.79KK270 pKa = 10.79EE271 pKa = 4.33AEE273 pKa = 4.02KK274 pKa = 10.9SRR276 pKa = 11.84EE277 pKa = 3.91SEE279 pKa = 3.29FDD281 pKa = 3.71YY282 pKa = 11.55NFGRR286 pKa = 11.84SNDD289 pKa = 3.48NEE291 pKa = 3.83FRR293 pKa = 11.84SCKK296 pKa = 9.93SLCAYY301 pKa = 7.86DD302 pKa = 3.83TTGPACGSDD311 pKa = 2.86GQTYY315 pKa = 10.86SNEE318 pKa = 3.61CMMRR322 pKa = 11.84CLEE325 pKa = 4.27PDD327 pKa = 2.52IKK329 pKa = 10.78YY330 pKa = 10.17YY331 pKa = 10.83HH332 pKa = 7.06EE333 pKa = 5.57GYY335 pKa = 10.43CLQQEE340 pKa = 4.46DD341 pKa = 4.47SSCDD345 pKa = 3.2VDD347 pKa = 4.87CSHH350 pKa = 6.65THH352 pKa = 7.18DD353 pKa = 4.76LVCGIDD359 pKa = 3.2GQTYY363 pKa = 10.5INYY366 pKa = 9.58CLYY369 pKa = 11.02AVTYY373 pKa = 9.84CDD375 pKa = 4.6KK376 pKa = 11.2NLATLPFLFGACEE389 pKa = 4.06SDD391 pKa = 3.56KK392 pKa = 10.71VTAFF396 pKa = 3.46
Molecular weight: 45.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A024GJV7|A0A024GJV7_9STRA Uncharacterized protein OS=Albugo candida OX=65357 GN=BN9_079480 PE=3 SV=1
MM1 pKa = 7.43IGHH4 pKa = 7.22RR5 pKa = 11.84DD6 pKa = 3.47SLRR9 pKa = 11.84DD10 pKa = 3.31AVIAKK15 pKa = 9.81KK16 pKa = 10.84VEE18 pKa = 4.21TVGIRR23 pKa = 11.84GDD25 pKa = 3.34VCNTSRR31 pKa = 11.84RR32 pKa = 11.84EE33 pKa = 3.76WRR35 pKa = 11.84TVTNLKK41 pKa = 10.17VSIMSKK47 pKa = 10.32SFAHH51 pKa = 6.62IDD53 pKa = 3.25AEE55 pKa = 4.16NRR57 pKa = 11.84QGALIIACEE66 pKa = 4.12PSTSFSGPYY75 pKa = 8.3QVCRR79 pKa = 11.84LSSTSEE85 pKa = 3.87STLVRR90 pKa = 11.84SVFLVVFKK98 pKa = 10.85KK99 pKa = 10.81AIGRR103 pKa = 11.84CEE105 pKa = 3.87EE106 pKa = 3.85HH107 pKa = 6.43CTRR110 pKa = 11.84GVFVLRR116 pKa = 11.84IRR118 pKa = 11.84YY119 pKa = 8.38HH120 pKa = 7.88VGIEE124 pKa = 3.96CCRR127 pKa = 11.84NGVRR131 pKa = 11.84TFVRR135 pKa = 11.84LIGLRR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 3.54
MM1 pKa = 7.43IGHH4 pKa = 7.22RR5 pKa = 11.84DD6 pKa = 3.47SLRR9 pKa = 11.84DD10 pKa = 3.31AVIAKK15 pKa = 9.81KK16 pKa = 10.84VEE18 pKa = 4.21TVGIRR23 pKa = 11.84GDD25 pKa = 3.34VCNTSRR31 pKa = 11.84RR32 pKa = 11.84EE33 pKa = 3.76WRR35 pKa = 11.84TVTNLKK41 pKa = 10.17VSIMSKK47 pKa = 10.32SFAHH51 pKa = 6.62IDD53 pKa = 3.25AEE55 pKa = 4.16NRR57 pKa = 11.84QGALIIACEE66 pKa = 4.12PSTSFSGPYY75 pKa = 8.3QVCRR79 pKa = 11.84LSSTSEE85 pKa = 3.87STLVRR90 pKa = 11.84SVFLVVFKK98 pKa = 10.85KK99 pKa = 10.81AIGRR103 pKa = 11.84CEE105 pKa = 3.87EE106 pKa = 3.85HH107 pKa = 6.43CTRR110 pKa = 11.84GVFVLRR116 pKa = 11.84IRR118 pKa = 11.84YY119 pKa = 8.38HH120 pKa = 7.88VGIEE124 pKa = 3.96CCRR127 pKa = 11.84NGVRR131 pKa = 11.84TFVRR135 pKa = 11.84LIGLRR140 pKa = 11.84RR141 pKa = 11.84RR142 pKa = 11.84RR143 pKa = 3.54
Molecular weight: 16.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6569959 |
49 |
14607 |
497.7 |
56.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.788 ± 0.015 | 2.073 ± 0.01 |
5.405 ± 0.013 | 6.622 ± 0.021 |
4.079 ± 0.013 | 4.813 ± 0.017 |
2.847 ± 0.01 | 5.97 ± 0.015 |
5.802 ± 0.023 | 9.68 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.438 ± 0.007 | 4.321 ± 0.011 |
3.953 ± 0.015 | 4.651 ± 0.014 |
6.018 ± 0.017 | 8.817 ± 0.023 |
5.688 ± 0.013 | 5.968 ± 0.014 |
1.146 ± 0.006 | 2.923 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
