
Candidatus Methanomethylophilus sp. 1R26
Taxonomy: cellular organisms; Archaea; Candidatus Thermoplasmatota; Thermoplasmata; Methanomassiliicoccales; Candidatus Methanomethylophilaceae; Candidatus Methanomethylophilus; unclassified Candidatus Methanomethylophilus
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
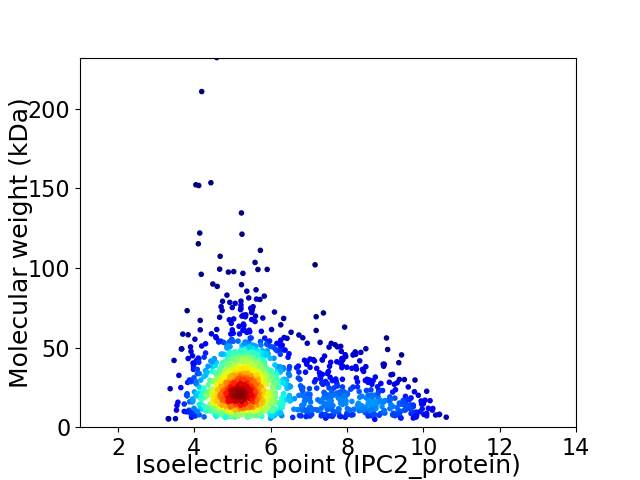
Virtual 2D-PAGE plot for 1473 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0W7THA7|A0A0W7THA7_9ARCH Phosphoesterase OS=Candidatus Methanomethylophilus sp. 1R26 OX=1769296 GN=AUQ37_03250 PE=4 SV=1
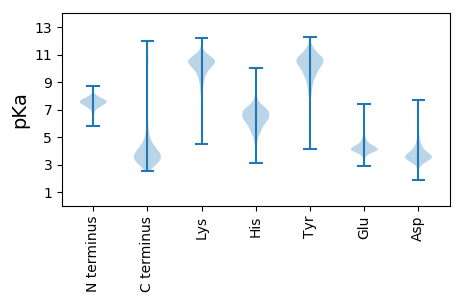
MM1 pKa = 7.9ADD3 pKa = 3.07QAVTYY8 pKa = 10.72GEE10 pKa = 4.13NTKK13 pKa = 10.8LSANAFSKK21 pKa = 10.66DD22 pKa = 3.27GYY24 pKa = 11.49SFGGWMFKK32 pKa = 10.85DD33 pKa = 3.56VVFADD38 pKa = 3.54EE39 pKa = 5.48AYY41 pKa = 9.77ILNLTSEE48 pKa = 4.7ADD50 pKa = 3.39AEE52 pKa = 4.46LTFTAVWIVNSYY64 pKa = 9.34TISFVSDD71 pKa = 3.65DD72 pKa = 4.09PVSGTTEE79 pKa = 3.76MQRR82 pKa = 11.84FTVEE86 pKa = 3.83DD87 pKa = 3.32EE88 pKa = 4.96GYY90 pKa = 8.48ATAVLSLNGFQRR102 pKa = 11.84AGYY105 pKa = 10.05SFAGWSDD112 pKa = 3.35GNRR115 pKa = 11.84TYY117 pKa = 11.57SDD119 pKa = 3.39GSLISTIDD127 pKa = 3.27LSKK130 pKa = 10.89AVSEE134 pKa = 4.84DD135 pKa = 3.37GVLTIALTAVWTANSYY151 pKa = 9.86TIFFDD156 pKa = 3.89TNGGSAIDD164 pKa = 5.3SIVADD169 pKa = 4.63CGTAIAVPEE178 pKa = 4.14PTKK181 pKa = 10.59EE182 pKa = 4.24GYY184 pKa = 8.11TFAGWLKK191 pKa = 10.88DD192 pKa = 3.18GAAYY196 pKa = 9.93SVPEE200 pKa = 4.11TMPAGDD206 pKa = 4.27LTLKK210 pKa = 10.79ASWKK214 pKa = 10.08LYY216 pKa = 10.1IPAPVNGVISLNAEE230 pKa = 3.63TDD232 pKa = 3.64GNLTLTDD239 pKa = 3.86EE240 pKa = 5.17SIGTLKK246 pKa = 10.67SYY248 pKa = 11.03ASSDD252 pKa = 3.66SAFLRR257 pKa = 11.84VSMGDD262 pKa = 3.15STFTFDD268 pKa = 3.97SKK270 pKa = 11.49AVAALNAGTLTIRR283 pKa = 11.84AMEE286 pKa = 4.32NLDD289 pKa = 4.55DD290 pKa = 3.85EE291 pKa = 4.86TAALTKK297 pKa = 10.5DD298 pKa = 3.09AAVYY302 pKa = 10.37AVGFGSNTYY311 pKa = 10.81LNGGTVTVTVPYY323 pKa = 9.1TLKK326 pKa = 10.77DD327 pKa = 3.52GQSADD332 pKa = 3.51DD333 pKa = 3.62VSIYY337 pKa = 10.24RR338 pKa = 11.84ISDD341 pKa = 3.28GSVAEE346 pKa = 4.5TIEE349 pKa = 4.05AQYY352 pKa = 11.53SDD354 pKa = 3.65GTVTFATGNLSSLYY368 pKa = 10.48AVGHH372 pKa = 6.98IDD374 pKa = 3.82TGSDD378 pKa = 4.1DD379 pKa = 4.92EE380 pKa = 6.01DD381 pKa = 5.41DD382 pKa = 3.87STAIMAVALAAVAVCLVAGIVYY404 pKa = 9.98AYY406 pKa = 9.94SRR408 pKa = 11.84RR409 pKa = 11.84VV410 pKa = 2.96
MM1 pKa = 7.9ADD3 pKa = 3.07QAVTYY8 pKa = 10.72GEE10 pKa = 4.13NTKK13 pKa = 10.8LSANAFSKK21 pKa = 10.66DD22 pKa = 3.27GYY24 pKa = 11.49SFGGWMFKK32 pKa = 10.85DD33 pKa = 3.56VVFADD38 pKa = 3.54EE39 pKa = 5.48AYY41 pKa = 9.77ILNLTSEE48 pKa = 4.7ADD50 pKa = 3.39AEE52 pKa = 4.46LTFTAVWIVNSYY64 pKa = 9.34TISFVSDD71 pKa = 3.65DD72 pKa = 4.09PVSGTTEE79 pKa = 3.76MQRR82 pKa = 11.84FTVEE86 pKa = 3.83DD87 pKa = 3.32EE88 pKa = 4.96GYY90 pKa = 8.48ATAVLSLNGFQRR102 pKa = 11.84AGYY105 pKa = 10.05SFAGWSDD112 pKa = 3.35GNRR115 pKa = 11.84TYY117 pKa = 11.57SDD119 pKa = 3.39GSLISTIDD127 pKa = 3.27LSKK130 pKa = 10.89AVSEE134 pKa = 4.84DD135 pKa = 3.37GVLTIALTAVWTANSYY151 pKa = 9.86TIFFDD156 pKa = 3.89TNGGSAIDD164 pKa = 5.3SIVADD169 pKa = 4.63CGTAIAVPEE178 pKa = 4.14PTKK181 pKa = 10.59EE182 pKa = 4.24GYY184 pKa = 8.11TFAGWLKK191 pKa = 10.88DD192 pKa = 3.18GAAYY196 pKa = 9.93SVPEE200 pKa = 4.11TMPAGDD206 pKa = 4.27LTLKK210 pKa = 10.79ASWKK214 pKa = 10.08LYY216 pKa = 10.1IPAPVNGVISLNAEE230 pKa = 3.63TDD232 pKa = 3.64GNLTLTDD239 pKa = 3.86EE240 pKa = 5.17SIGTLKK246 pKa = 10.67SYY248 pKa = 11.03ASSDD252 pKa = 3.66SAFLRR257 pKa = 11.84VSMGDD262 pKa = 3.15STFTFDD268 pKa = 3.97SKK270 pKa = 11.49AVAALNAGTLTIRR283 pKa = 11.84AMEE286 pKa = 4.32NLDD289 pKa = 4.55DD290 pKa = 3.85EE291 pKa = 4.86TAALTKK297 pKa = 10.5DD298 pKa = 3.09AAVYY302 pKa = 10.37AVGFGSNTYY311 pKa = 10.81LNGGTVTVTVPYY323 pKa = 9.1TLKK326 pKa = 10.77DD327 pKa = 3.52GQSADD332 pKa = 3.51DD333 pKa = 3.62VSIYY337 pKa = 10.24RR338 pKa = 11.84ISDD341 pKa = 3.28GSVAEE346 pKa = 4.5TIEE349 pKa = 4.05AQYY352 pKa = 11.53SDD354 pKa = 3.65GTVTFATGNLSSLYY368 pKa = 10.48AVGHH372 pKa = 6.98IDD374 pKa = 3.82TGSDD378 pKa = 4.1DD379 pKa = 4.92EE380 pKa = 6.01DD381 pKa = 5.41DD382 pKa = 3.87STAIMAVALAAVAVCLVAGIVYY404 pKa = 9.98AYY406 pKa = 9.94SRR408 pKa = 11.84RR409 pKa = 11.84VV410 pKa = 2.96
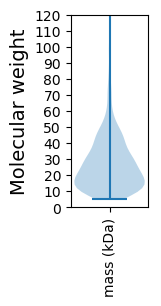
Molecular weight: 43.14 kDa
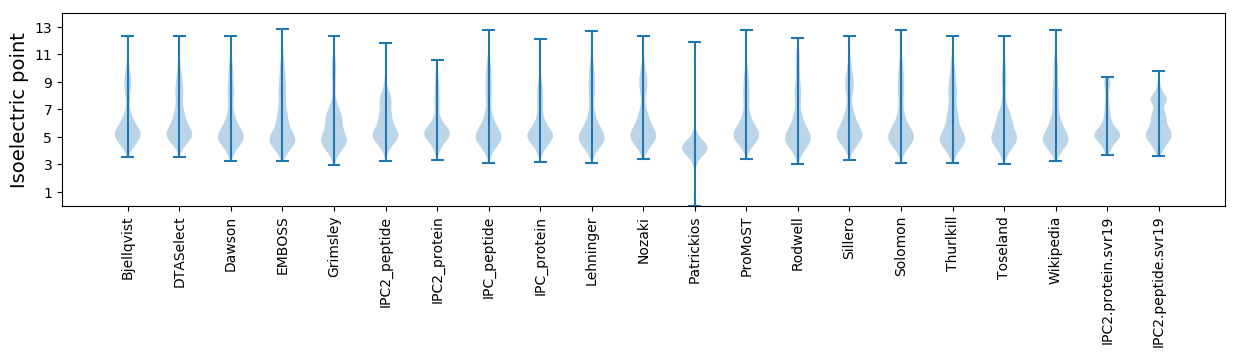
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0W7TJX8|A0A0W7TJX8_9ARCH Protein translation factor SUI1 homolog OS=Candidatus Methanomethylophilus sp. 1R26 OX=1769296 GN=AUQ37_05690 PE=3 SV=1
MM1 pKa = 7.46KK2 pKa = 9.88PKK4 pKa = 10.03KK5 pKa = 10.17QYY7 pKa = 10.21GRR9 pKa = 11.84QVGCDD14 pKa = 2.55RR15 pKa = 11.84CGRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84GIIRR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 10.09GMHH29 pKa = 7.66LCRR32 pKa = 11.84QCFRR36 pKa = 11.84DD37 pKa = 3.69MAPEE41 pKa = 4.41LGFKK45 pKa = 10.27KK46 pKa = 10.81YY47 pKa = 10.16SS48 pKa = 3.24
MM1 pKa = 7.46KK2 pKa = 9.88PKK4 pKa = 10.03KK5 pKa = 10.17QYY7 pKa = 10.21GRR9 pKa = 11.84QVGCDD14 pKa = 2.55RR15 pKa = 11.84CGRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84GIIRR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 10.09GMHH29 pKa = 7.66LCRR32 pKa = 11.84QCFRR36 pKa = 11.84DD37 pKa = 3.69MAPEE41 pKa = 4.41LGFKK45 pKa = 10.27KK46 pKa = 10.81YY47 pKa = 10.16SS48 pKa = 3.24
Molecular weight: 5.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
376912 |
43 |
2263 |
255.9 |
28.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.548 ± 0.091 | 1.744 ± 0.034 |
6.619 ± 0.054 | 6.455 ± 0.073 |
3.661 ± 0.046 | 8.447 ± 0.065 |
1.535 ± 0.026 | 6.309 ± 0.059 |
5.049 ± 0.065 | 7.907 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.326 ± 0.046 | 2.996 ± 0.045 |
4.145 ± 0.044 | 2.093 ± 0.032 |
5.965 ± 0.085 | 7.106 ± 0.093 |
5.273 ± 0.084 | 7.318 ± 0.056 |
1.039 ± 0.025 | 3.465 ± 0.046 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
