
Arthrobacter phage Kuleana
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Kuleanavirus; Arthrobacter virus Kuleana
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
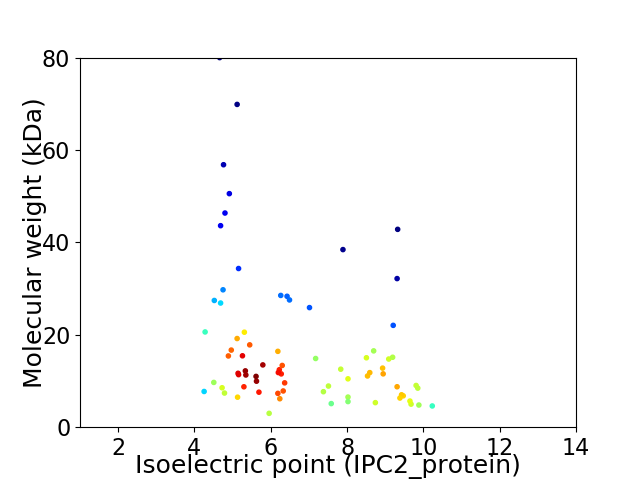
Virtual 2D-PAGE plot for 76 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
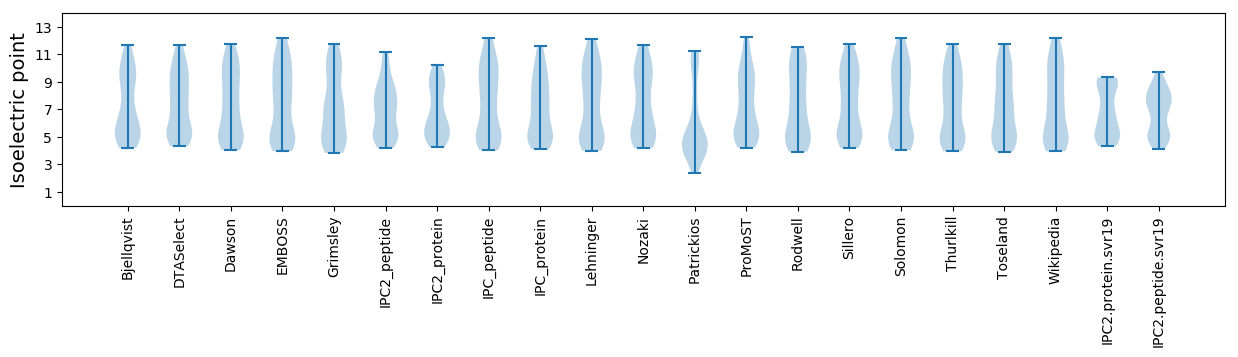
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5Q2W8L7|A0A5Q2W8L7_9CAUD RusA-like resolvase OS=Arthrobacter phage Kuleana OX=2653270 GN=45 PE=4 SV=1
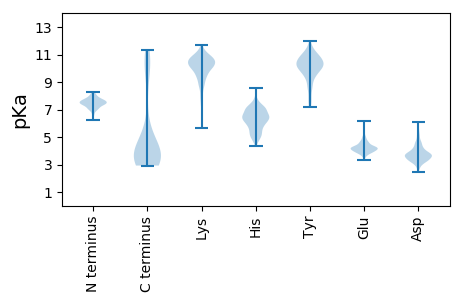
MM1 pKa = 7.5KK2 pKa = 10.49LVTINGNGGNDD13 pKa = 3.4YY14 pKa = 10.98DD15 pKa = 4.62VSVHH19 pKa = 6.7RR20 pKa = 11.84ADD22 pKa = 4.4CADD25 pKa = 3.39VPKK28 pKa = 9.97ATRR31 pKa = 11.84GHH33 pKa = 6.97DD34 pKa = 3.64YY35 pKa = 10.86FEE37 pKa = 5.06EE38 pKa = 4.02EE39 pKa = 4.5HH40 pKa = 5.95EE41 pKa = 4.63SKK43 pKa = 10.63RR44 pKa = 11.84ALWLDD49 pKa = 3.52YY50 pKa = 11.14NADD53 pKa = 3.78FLAEE57 pKa = 4.54GPGSAWPIHH66 pKa = 6.48FYY68 pKa = 10.59PCTAGLPDD76 pKa = 4.09GGTYY80 pKa = 8.78NTDD83 pKa = 3.17TDD85 pKa = 3.57EE86 pKa = 4.64GKK88 pKa = 10.61
MM1 pKa = 7.5KK2 pKa = 10.49LVTINGNGGNDD13 pKa = 3.4YY14 pKa = 10.98DD15 pKa = 4.62VSVHH19 pKa = 6.7RR20 pKa = 11.84ADD22 pKa = 4.4CADD25 pKa = 3.39VPKK28 pKa = 9.97ATRR31 pKa = 11.84GHH33 pKa = 6.97DD34 pKa = 3.64YY35 pKa = 10.86FEE37 pKa = 5.06EE38 pKa = 4.02EE39 pKa = 4.5HH40 pKa = 5.95EE41 pKa = 4.63SKK43 pKa = 10.63RR44 pKa = 11.84ALWLDD49 pKa = 3.52YY50 pKa = 11.14NADD53 pKa = 3.78FLAEE57 pKa = 4.54GPGSAWPIHH66 pKa = 6.48FYY68 pKa = 10.59PCTAGLPDD76 pKa = 4.09GGTYY80 pKa = 8.78NTDD83 pKa = 3.17TDD85 pKa = 3.57EE86 pKa = 4.64GKK88 pKa = 10.61
Molecular weight: 9.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2WE41|A0A5Q2WE41_9CAUD Uncharacterized protein OS=Arthrobacter phage Kuleana OX=2653270 GN=33 PE=4 SV=1
MM1 pKa = 7.52GRR3 pKa = 11.84PPLPVGTWGAIRR15 pKa = 11.84VEE17 pKa = 4.82RR18 pKa = 11.84IAGGYY23 pKa = 7.67RR24 pKa = 11.84ARR26 pKa = 11.84ARR28 pKa = 11.84FRR30 pKa = 11.84DD31 pKa = 3.49FDD33 pKa = 3.39GRR35 pKa = 11.84TRR37 pKa = 11.84DD38 pKa = 3.38VEE40 pKa = 4.21RR41 pKa = 11.84SGKK44 pKa = 8.31TKK46 pKa = 10.24GAARR50 pKa = 11.84AALTADD56 pKa = 3.52LTEE59 pKa = 4.3RR60 pKa = 11.84TAPAGDD66 pKa = 4.84EE67 pKa = 4.08ITASTRR73 pKa = 11.84LQLVAEE79 pKa = 4.83IWRR82 pKa = 11.84TEE84 pKa = 3.57KK85 pKa = 10.55WPDD88 pKa = 3.01LAEE91 pKa = 4.31NSRR94 pKa = 11.84KK95 pKa = 9.83RR96 pKa = 11.84YY97 pKa = 9.47RR98 pKa = 11.84DD99 pKa = 3.27ALEE102 pKa = 4.26DD103 pKa = 4.42HH104 pKa = 6.97ILPGLGALTVSEE116 pKa = 4.59CSVTRR121 pKa = 11.84IDD123 pKa = 4.51RR124 pKa = 11.84FLKK127 pKa = 9.48ATAASTGAPSAKK139 pKa = 9.33VCRR142 pKa = 11.84SVLSGILGLAVRR154 pKa = 11.84HH155 pKa = 5.79GAAATNPVRR164 pKa = 11.84DD165 pKa = 3.64VAGITVTPKK174 pKa = 10.38EE175 pKa = 4.0IRR177 pKa = 11.84ALTLEE182 pKa = 4.66EE183 pKa = 3.61IRR185 pKa = 11.84AARR188 pKa = 11.84AAVRR192 pKa = 11.84SWQLGEE198 pKa = 4.0PLAEE202 pKa = 4.04GRR204 pKa = 11.84PRR206 pKa = 11.84RR207 pKa = 11.84GRR209 pKa = 11.84PPTQDD214 pKa = 3.83LLDD217 pKa = 4.06ILDD220 pKa = 4.63LLLATGARR228 pKa = 11.84IGEE231 pKa = 4.28LLAVQWSDD239 pKa = 3.16VDD241 pKa = 5.71LEE243 pKa = 4.3AQTLTISGTIISTEE257 pKa = 4.08EE258 pKa = 3.79KK259 pKa = 9.53PSRR262 pKa = 11.84LVRR265 pKa = 11.84QAHH268 pKa = 6.22PKK270 pKa = 9.42SSSSRR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84LTLPPFAIDD286 pKa = 2.95ALMRR290 pKa = 11.84RR291 pKa = 11.84RR292 pKa = 11.84LAVTVANVHH301 pKa = 6.45DD302 pKa = 4.25VVFPSTEE309 pKa = 3.7GTLRR313 pKa = 11.84DD314 pKa = 3.73PGSVRR319 pKa = 11.84KK320 pKa = 9.05QLGKK324 pKa = 10.57VLGPAGLGWVTPHH337 pKa = 6.16VFRR340 pKa = 11.84KK341 pKa = 7.9TVATVLDD348 pKa = 3.78RR349 pKa = 11.84AEE351 pKa = 4.51DD352 pKa = 3.53LRR354 pKa = 11.84TASDD358 pKa = 3.67QLGHH362 pKa = 7.23AGTDD366 pKa = 3.2VTRR369 pKa = 11.84RR370 pKa = 11.84HH371 pKa = 4.93YY372 pKa = 10.54VEE374 pKa = 4.53RR375 pKa = 11.84THH377 pKa = 6.64EE378 pKa = 4.5GPDD381 pKa = 3.15VRR383 pKa = 11.84EE384 pKa = 3.8ALEE387 pKa = 3.88RR388 pKa = 11.84LVRR391 pKa = 11.84GSS393 pKa = 3.1
MM1 pKa = 7.52GRR3 pKa = 11.84PPLPVGTWGAIRR15 pKa = 11.84VEE17 pKa = 4.82RR18 pKa = 11.84IAGGYY23 pKa = 7.67RR24 pKa = 11.84ARR26 pKa = 11.84ARR28 pKa = 11.84FRR30 pKa = 11.84DD31 pKa = 3.49FDD33 pKa = 3.39GRR35 pKa = 11.84TRR37 pKa = 11.84DD38 pKa = 3.38VEE40 pKa = 4.21RR41 pKa = 11.84SGKK44 pKa = 8.31TKK46 pKa = 10.24GAARR50 pKa = 11.84AALTADD56 pKa = 3.52LTEE59 pKa = 4.3RR60 pKa = 11.84TAPAGDD66 pKa = 4.84EE67 pKa = 4.08ITASTRR73 pKa = 11.84LQLVAEE79 pKa = 4.83IWRR82 pKa = 11.84TEE84 pKa = 3.57KK85 pKa = 10.55WPDD88 pKa = 3.01LAEE91 pKa = 4.31NSRR94 pKa = 11.84KK95 pKa = 9.83RR96 pKa = 11.84YY97 pKa = 9.47RR98 pKa = 11.84DD99 pKa = 3.27ALEE102 pKa = 4.26DD103 pKa = 4.42HH104 pKa = 6.97ILPGLGALTVSEE116 pKa = 4.59CSVTRR121 pKa = 11.84IDD123 pKa = 4.51RR124 pKa = 11.84FLKK127 pKa = 9.48ATAASTGAPSAKK139 pKa = 9.33VCRR142 pKa = 11.84SVLSGILGLAVRR154 pKa = 11.84HH155 pKa = 5.79GAAATNPVRR164 pKa = 11.84DD165 pKa = 3.64VAGITVTPKK174 pKa = 10.38EE175 pKa = 4.0IRR177 pKa = 11.84ALTLEE182 pKa = 4.66EE183 pKa = 3.61IRR185 pKa = 11.84AARR188 pKa = 11.84AAVRR192 pKa = 11.84SWQLGEE198 pKa = 4.0PLAEE202 pKa = 4.04GRR204 pKa = 11.84PRR206 pKa = 11.84RR207 pKa = 11.84GRR209 pKa = 11.84PPTQDD214 pKa = 3.83LLDD217 pKa = 4.06ILDD220 pKa = 4.63LLLATGARR228 pKa = 11.84IGEE231 pKa = 4.28LLAVQWSDD239 pKa = 3.16VDD241 pKa = 5.71LEE243 pKa = 4.3AQTLTISGTIISTEE257 pKa = 4.08EE258 pKa = 3.79KK259 pKa = 9.53PSRR262 pKa = 11.84LVRR265 pKa = 11.84QAHH268 pKa = 6.22PKK270 pKa = 9.42SSSSRR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84LTLPPFAIDD286 pKa = 2.95ALMRR290 pKa = 11.84RR291 pKa = 11.84RR292 pKa = 11.84LAVTVANVHH301 pKa = 6.45DD302 pKa = 4.25VVFPSTEE309 pKa = 3.7GTLRR313 pKa = 11.84DD314 pKa = 3.73PGSVRR319 pKa = 11.84KK320 pKa = 9.05QLGKK324 pKa = 10.57VLGPAGLGWVTPHH337 pKa = 6.16VFRR340 pKa = 11.84KK341 pKa = 7.9TVATVLDD348 pKa = 3.78RR349 pKa = 11.84AEE351 pKa = 4.51DD352 pKa = 3.53LRR354 pKa = 11.84TASDD358 pKa = 3.67QLGHH362 pKa = 7.23AGTDD366 pKa = 3.2VTRR369 pKa = 11.84RR370 pKa = 11.84HH371 pKa = 4.93YY372 pKa = 10.54VEE374 pKa = 4.53RR375 pKa = 11.84THH377 pKa = 6.64EE378 pKa = 4.5GPDD381 pKa = 3.15VRR383 pKa = 11.84EE384 pKa = 3.8ALEE387 pKa = 3.88RR388 pKa = 11.84LVRR391 pKa = 11.84GSS393 pKa = 3.1
Molecular weight: 42.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12284 |
26 |
787 |
161.6 |
17.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.61 ± 0.385 | 0.944 ± 0.121 |
5.698 ± 0.194 | 6.472 ± 0.35 |
2.613 ± 0.225 | 9.964 ± 0.535 |
1.53 ± 0.144 | 3.623 ± 0.206 |
3.598 ± 0.26 | 9.02 ± 0.249 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.043 ± 0.114 | 2.369 ± 0.182 |
5.536 ± 0.319 | 2.703 ± 0.172 |
7.766 ± 0.413 | 5.332 ± 0.301 |
5.528 ± 0.334 | 8.344 ± 0.368 |
2.361 ± 0.156 | 1.946 ± 0.142 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
