
Cherry associated luteovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Luteovirus
Average proteome isoelectric point is 7.87
Get precalculated fractions of proteins
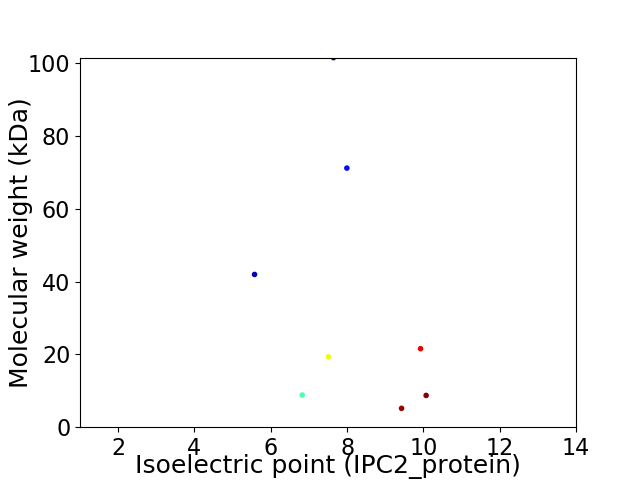
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I9W765|A0A1I9W765_9LUTE RNA-directed RNA polymerase OS=Cherry associated luteovirus OX=1912598 PE=4 SV=1
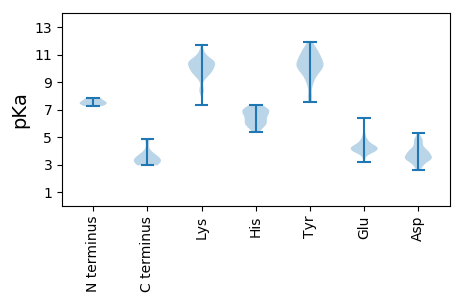
MM1 pKa = 7.57LFDD4 pKa = 3.88TLLFASAKK12 pKa = 9.36LVKK15 pKa = 10.71DD16 pKa = 4.14FVSYY20 pKa = 10.47LYY22 pKa = 11.25ANLKK26 pKa = 9.79FVYY29 pKa = 9.91KK30 pKa = 10.29SLKK33 pKa = 9.55RR34 pKa = 11.84WLLEE38 pKa = 3.72LQGKK42 pKa = 8.25FRR44 pKa = 11.84QHH46 pKa = 6.93DD47 pKa = 3.87SFVDD51 pKa = 3.53MVYY54 pKa = 11.22GFMDD58 pKa = 3.76DD59 pKa = 4.39VEE61 pKa = 5.55DD62 pKa = 5.0FEE64 pKa = 4.84FDD66 pKa = 3.86CVVDD70 pKa = 4.95LEE72 pKa = 4.2AAEE75 pKa = 4.34LQFSKK80 pKa = 10.79AKK82 pKa = 10.63YY83 pKa = 9.17MLEE86 pKa = 3.9QMEE89 pKa = 4.6KK90 pKa = 10.49ARR92 pKa = 11.84GVLPEE97 pKa = 4.29WPQPMRR103 pKa = 11.84PLGADD108 pKa = 3.66PIGPPTLIDD117 pKa = 3.75GEE119 pKa = 4.71VIPDD123 pKa = 4.7PIVDD127 pKa = 3.35RR128 pKa = 11.84VKK130 pKa = 10.98FEE132 pKa = 3.91NDD134 pKa = 2.8QAHH137 pKa = 6.48VIVQNAFSEE146 pKa = 4.55AEE148 pKa = 3.92ILGQTSIFCHH158 pKa = 5.95AKK160 pKa = 9.71YY161 pKa = 10.76DD162 pKa = 4.61LIQAEE167 pKa = 3.95EE168 pKa = 4.4RR169 pKa = 11.84IRR171 pKa = 11.84DD172 pKa = 3.25KK173 pKa = 11.11YY174 pKa = 10.88ARR176 pKa = 11.84EE177 pKa = 3.88KK178 pKa = 10.56GEE180 pKa = 4.08HH181 pKa = 5.39YY182 pKa = 10.39FGRR185 pKa = 11.84FINTFSSRR193 pKa = 11.84LAYY196 pKa = 9.99VKK198 pKa = 10.25KK199 pKa = 10.1CAARR203 pKa = 11.84RR204 pKa = 11.84IEE206 pKa = 4.03SSRR209 pKa = 11.84LAEE212 pKa = 4.08KK213 pKa = 10.19VQKK216 pKa = 10.41QVFQVDD222 pKa = 3.56NVPDD226 pKa = 4.78FEE228 pKa = 5.24AMCDD232 pKa = 3.34MVQIEE237 pKa = 4.23TGEE240 pKa = 4.19TKK242 pKa = 9.81EE243 pKa = 4.18VKK245 pKa = 10.42GGEE248 pKa = 4.32DD249 pKa = 3.42GSEE252 pKa = 3.96PTTVPIKK259 pKa = 10.11KK260 pKa = 8.49WVRR263 pKa = 11.84RR264 pKa = 11.84LRR266 pKa = 11.84LDD268 pKa = 3.43DD269 pKa = 4.79AKK271 pKa = 11.19VKK273 pKa = 10.57KK274 pKa = 9.85DD275 pKa = 2.81ACKK278 pKa = 10.12FIRR281 pKa = 11.84RR282 pKa = 11.84YY283 pKa = 9.89VIHH286 pKa = 6.83HH287 pKa = 5.68NCRR290 pKa = 11.84LNSDD294 pKa = 4.02EE295 pKa = 4.32VSVMTINRR303 pKa = 11.84YY304 pKa = 8.03VAQFCDD310 pKa = 5.29LFKK313 pKa = 10.96LDD315 pKa = 4.54QNSTDD320 pKa = 3.95YY321 pKa = 11.59LMKK324 pKa = 10.53AALLMVPIVTKK335 pKa = 10.47EE336 pKa = 3.97DD337 pKa = 3.59LVSTMVIHH345 pKa = 6.98SPAARR350 pKa = 11.84DD351 pKa = 3.43LRR353 pKa = 11.84AMKK356 pKa = 9.38DD357 pKa = 3.43TVEE360 pKa = 4.28SGVFF364 pKa = 3.19
MM1 pKa = 7.57LFDD4 pKa = 3.88TLLFASAKK12 pKa = 9.36LVKK15 pKa = 10.71DD16 pKa = 4.14FVSYY20 pKa = 10.47LYY22 pKa = 11.25ANLKK26 pKa = 9.79FVYY29 pKa = 9.91KK30 pKa = 10.29SLKK33 pKa = 9.55RR34 pKa = 11.84WLLEE38 pKa = 3.72LQGKK42 pKa = 8.25FRR44 pKa = 11.84QHH46 pKa = 6.93DD47 pKa = 3.87SFVDD51 pKa = 3.53MVYY54 pKa = 11.22GFMDD58 pKa = 3.76DD59 pKa = 4.39VEE61 pKa = 5.55DD62 pKa = 5.0FEE64 pKa = 4.84FDD66 pKa = 3.86CVVDD70 pKa = 4.95LEE72 pKa = 4.2AAEE75 pKa = 4.34LQFSKK80 pKa = 10.79AKK82 pKa = 10.63YY83 pKa = 9.17MLEE86 pKa = 3.9QMEE89 pKa = 4.6KK90 pKa = 10.49ARR92 pKa = 11.84GVLPEE97 pKa = 4.29WPQPMRR103 pKa = 11.84PLGADD108 pKa = 3.66PIGPPTLIDD117 pKa = 3.75GEE119 pKa = 4.71VIPDD123 pKa = 4.7PIVDD127 pKa = 3.35RR128 pKa = 11.84VKK130 pKa = 10.98FEE132 pKa = 3.91NDD134 pKa = 2.8QAHH137 pKa = 6.48VIVQNAFSEE146 pKa = 4.55AEE148 pKa = 3.92ILGQTSIFCHH158 pKa = 5.95AKK160 pKa = 9.71YY161 pKa = 10.76DD162 pKa = 4.61LIQAEE167 pKa = 3.95EE168 pKa = 4.4RR169 pKa = 11.84IRR171 pKa = 11.84DD172 pKa = 3.25KK173 pKa = 11.11YY174 pKa = 10.88ARR176 pKa = 11.84EE177 pKa = 3.88KK178 pKa = 10.56GEE180 pKa = 4.08HH181 pKa = 5.39YY182 pKa = 10.39FGRR185 pKa = 11.84FINTFSSRR193 pKa = 11.84LAYY196 pKa = 9.99VKK198 pKa = 10.25KK199 pKa = 10.1CAARR203 pKa = 11.84RR204 pKa = 11.84IEE206 pKa = 4.03SSRR209 pKa = 11.84LAEE212 pKa = 4.08KK213 pKa = 10.19VQKK216 pKa = 10.41QVFQVDD222 pKa = 3.56NVPDD226 pKa = 4.78FEE228 pKa = 5.24AMCDD232 pKa = 3.34MVQIEE237 pKa = 4.23TGEE240 pKa = 4.19TKK242 pKa = 9.81EE243 pKa = 4.18VKK245 pKa = 10.42GGEE248 pKa = 4.32DD249 pKa = 3.42GSEE252 pKa = 3.96PTTVPIKK259 pKa = 10.11KK260 pKa = 8.49WVRR263 pKa = 11.84RR264 pKa = 11.84LRR266 pKa = 11.84LDD268 pKa = 3.43DD269 pKa = 4.79AKK271 pKa = 11.19VKK273 pKa = 10.57KK274 pKa = 9.85DD275 pKa = 2.81ACKK278 pKa = 10.12FIRR281 pKa = 11.84RR282 pKa = 11.84YY283 pKa = 9.89VIHH286 pKa = 6.83HH287 pKa = 5.68NCRR290 pKa = 11.84LNSDD294 pKa = 4.02EE295 pKa = 4.32VSVMTINRR303 pKa = 11.84YY304 pKa = 8.03VAQFCDD310 pKa = 5.29LFKK313 pKa = 10.96LDD315 pKa = 4.54QNSTDD320 pKa = 3.95YY321 pKa = 11.59LMKK324 pKa = 10.53AALLMVPIVTKK335 pKa = 10.47EE336 pKa = 3.97DD337 pKa = 3.59LVSTMVIHH345 pKa = 6.98SPAARR350 pKa = 11.84DD351 pKa = 3.43LRR353 pKa = 11.84AMKK356 pKa = 9.38DD357 pKa = 3.43TVEE360 pKa = 4.28SGVFF364 pKa = 3.19
Molecular weight: 41.96 kDa
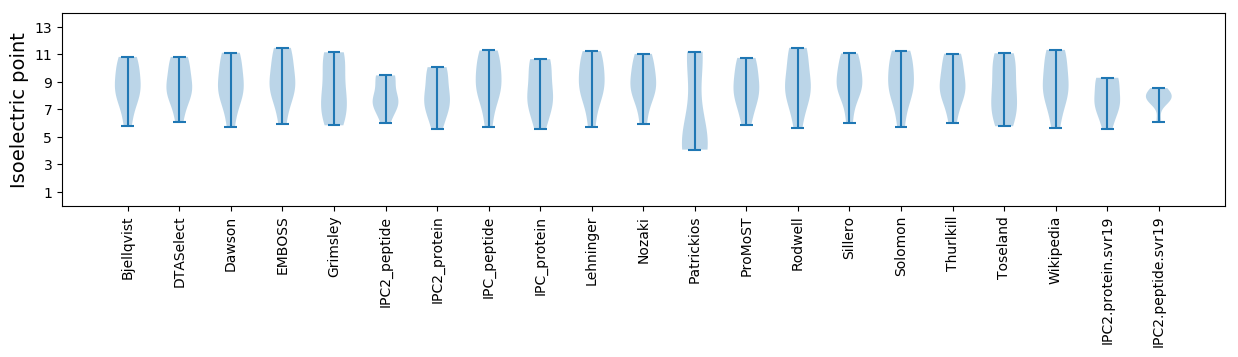
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I9W771|A0A1I9W771_9LUTE Putative P7 protein OS=Cherry associated luteovirus OX=1912598 PE=4 SV=1
MM1 pKa = 7.31NPGKK5 pKa = 9.77QVHH8 pKa = 5.99LVNSGVYY15 pKa = 9.58IGALIGGKK23 pKa = 9.89LGGSSTPPYY32 pKa = 10.1YY33 pKa = 10.42FWLVRR38 pKa = 11.84QTNSTKK44 pKa = 9.91IQKK47 pKa = 9.9QSVLLLSFMKK57 pKa = 10.25LRR59 pKa = 11.84SPIHH63 pKa = 6.18VGVKK67 pKa = 10.07NMGRR71 pKa = 11.84VEE73 pKa = 4.63LSQLCRR79 pKa = 3.49
MM1 pKa = 7.31NPGKK5 pKa = 9.77QVHH8 pKa = 5.99LVNSGVYY15 pKa = 9.58IGALIGGKK23 pKa = 9.89LGGSSTPPYY32 pKa = 10.1YY33 pKa = 10.42FWLVRR38 pKa = 11.84QTNSTKK44 pKa = 9.91IQKK47 pKa = 9.9QSVLLLSFMKK57 pKa = 10.25LRR59 pKa = 11.84SPIHH63 pKa = 6.18VGVKK67 pKa = 10.07NMGRR71 pKa = 11.84VEE73 pKa = 4.63LSQLCRR79 pKa = 3.49
Molecular weight: 8.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2477 |
45 |
890 |
309.6 |
34.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.773 ± 0.421 | 1.494 ± 0.335 |
5.612 ± 0.79 | 5.491 ± 0.675 |
5.127 ± 0.661 | 6.742 ± 1.136 |
1.655 ± 0.256 | 5.45 ± 0.234 |
6.5 ± 0.618 | 7.751 ± 0.929 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.584 ± 0.495 | 4.037 ± 0.705 |
5.531 ± 0.625 | 4.522 ± 0.375 |
6.419 ± 0.311 | 8.236 ± 1.399 |
4.723 ± 0.636 | 7.792 ± 0.824 |
1.171 ± 0.136 | 3.149 ± 0.225 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
