
Phlebiopsis gigantea 11061_1 CR5-6
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Phanerochaetaceae; Phlebiopsis; Phlebiopsis gigantea
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
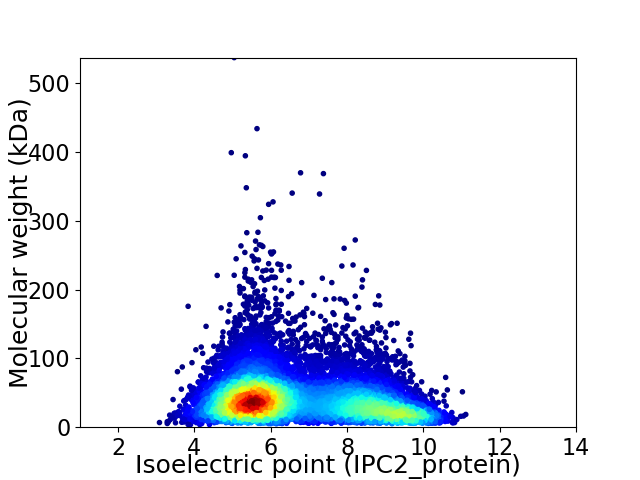
Virtual 2D-PAGE plot for 11848 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C3PGL3|A0A0C3PGL3_PHLGI Uncharacterized protein OS=Phlebiopsis gigantea 11061_1 CR5-6 OX=745531 GN=PHLGIDRAFT_129223 PE=4 SV=1
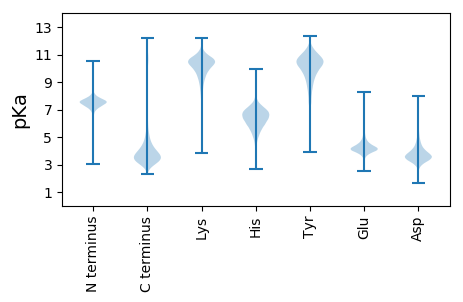
MM1 pKa = 6.82STEE4 pKa = 3.99PQSLFSLARR13 pKa = 11.84NKK15 pKa = 10.16LQSVVGGGVKK25 pKa = 10.32DD26 pKa = 3.46SSSLHH31 pKa = 5.33RR32 pKa = 11.84WVLLKK37 pKa = 10.98NSIIRR42 pKa = 11.84SHH44 pKa = 5.87VQTPATSQALDD55 pKa = 3.46EE56 pKa = 4.96AEE58 pKa = 4.12ADD60 pKa = 3.46SVYY63 pKa = 10.83RR64 pKa = 11.84QDD66 pKa = 3.4EE67 pKa = 4.32EE68 pKa = 4.55IYY70 pKa = 10.47EE71 pKa = 4.21VEE73 pKa = 4.02HH74 pKa = 6.86DD75 pKa = 4.23AFMFPDD81 pKa = 3.83PHH83 pKa = 6.44SACADD88 pKa = 3.26SSAAGNDD95 pKa = 3.21EE96 pKa = 4.09NQWLDD101 pKa = 3.45SVLEE105 pKa = 3.99GLGDD109 pKa = 4.47DD110 pKa = 4.12EE111 pKa = 7.28DD112 pKa = 6.37DD113 pKa = 3.97YY114 pKa = 11.62FQAGALPFADD124 pKa = 5.5DD125 pKa = 5.27DD126 pKa = 4.38IEE128 pKa = 4.53PTSPMYY134 pKa = 11.12SPMSSSDD141 pKa = 3.89DD142 pKa = 3.68LAEE145 pKa = 3.97QPSYY149 pKa = 11.35YY150 pKa = 9.83DD151 pKa = 3.33YY152 pKa = 11.37PLIYY156 pKa = 10.11PPFRR160 pKa = 11.84PPVPSTWSPLEE171 pKa = 4.15HH172 pKa = 6.89SSDD175 pKa = 4.06SILPSTSALYY185 pKa = 10.4DD186 pKa = 3.4NPLPYY191 pKa = 10.41LDD193 pKa = 4.85VDD195 pKa = 4.26EE196 pKa = 6.38LDD198 pKa = 4.65DD199 pKa = 5.25SPVPDD204 pKa = 5.51AIEE207 pKa = 4.38DD208 pKa = 3.94TSDD211 pKa = 4.1DD212 pKa = 3.89EE213 pKa = 5.66SDD215 pKa = 3.84APSTPSAVSTPSLSPPGPTIPMPRR239 pKa = 11.84EE240 pKa = 3.57RR241 pKa = 11.84TRR243 pKa = 11.84LLTHH247 pKa = 5.69PHH249 pKa = 5.99VYY251 pKa = 10.05VEE253 pKa = 4.13RR254 pKa = 11.84DD255 pKa = 3.18DD256 pKa = 4.77PYY258 pKa = 10.99FYY260 pKa = 10.53PFEE263 pKa = 6.33LDD265 pKa = 4.54LDD267 pKa = 4.32PLPLPDD273 pKa = 4.38DD274 pKa = 3.75TLAPSLRR281 pKa = 11.84AYY283 pKa = 10.37RR284 pKa = 11.84DD285 pKa = 3.7AYY287 pKa = 11.11SSEE290 pKa = 4.32CC291 pKa = 3.96
MM1 pKa = 6.82STEE4 pKa = 3.99PQSLFSLARR13 pKa = 11.84NKK15 pKa = 10.16LQSVVGGGVKK25 pKa = 10.32DD26 pKa = 3.46SSSLHH31 pKa = 5.33RR32 pKa = 11.84WVLLKK37 pKa = 10.98NSIIRR42 pKa = 11.84SHH44 pKa = 5.87VQTPATSQALDD55 pKa = 3.46EE56 pKa = 4.96AEE58 pKa = 4.12ADD60 pKa = 3.46SVYY63 pKa = 10.83RR64 pKa = 11.84QDD66 pKa = 3.4EE67 pKa = 4.32EE68 pKa = 4.55IYY70 pKa = 10.47EE71 pKa = 4.21VEE73 pKa = 4.02HH74 pKa = 6.86DD75 pKa = 4.23AFMFPDD81 pKa = 3.83PHH83 pKa = 6.44SACADD88 pKa = 3.26SSAAGNDD95 pKa = 3.21EE96 pKa = 4.09NQWLDD101 pKa = 3.45SVLEE105 pKa = 3.99GLGDD109 pKa = 4.47DD110 pKa = 4.12EE111 pKa = 7.28DD112 pKa = 6.37DD113 pKa = 3.97YY114 pKa = 11.62FQAGALPFADD124 pKa = 5.5DD125 pKa = 5.27DD126 pKa = 4.38IEE128 pKa = 4.53PTSPMYY134 pKa = 11.12SPMSSSDD141 pKa = 3.89DD142 pKa = 3.68LAEE145 pKa = 3.97QPSYY149 pKa = 11.35YY150 pKa = 9.83DD151 pKa = 3.33YY152 pKa = 11.37PLIYY156 pKa = 10.11PPFRR160 pKa = 11.84PPVPSTWSPLEE171 pKa = 4.15HH172 pKa = 6.89SSDD175 pKa = 4.06SILPSTSALYY185 pKa = 10.4DD186 pKa = 3.4NPLPYY191 pKa = 10.41LDD193 pKa = 4.85VDD195 pKa = 4.26EE196 pKa = 6.38LDD198 pKa = 4.65DD199 pKa = 5.25SPVPDD204 pKa = 5.51AIEE207 pKa = 4.38DD208 pKa = 3.94TSDD211 pKa = 4.1DD212 pKa = 3.89EE213 pKa = 5.66SDD215 pKa = 3.84APSTPSAVSTPSLSPPGPTIPMPRR239 pKa = 11.84EE240 pKa = 3.57RR241 pKa = 11.84TRR243 pKa = 11.84LLTHH247 pKa = 5.69PHH249 pKa = 5.99VYY251 pKa = 10.05VEE253 pKa = 4.13RR254 pKa = 11.84DD255 pKa = 3.18DD256 pKa = 4.77PYY258 pKa = 10.99FYY260 pKa = 10.53PFEE263 pKa = 6.33LDD265 pKa = 4.54LDD267 pKa = 4.32PLPLPDD273 pKa = 4.38DD274 pKa = 3.75TLAPSLRR281 pKa = 11.84AYY283 pKa = 10.37RR284 pKa = 11.84DD285 pKa = 3.7AYY287 pKa = 11.11SSEE290 pKa = 4.32CC291 pKa = 3.96
Molecular weight: 32.18 kDa
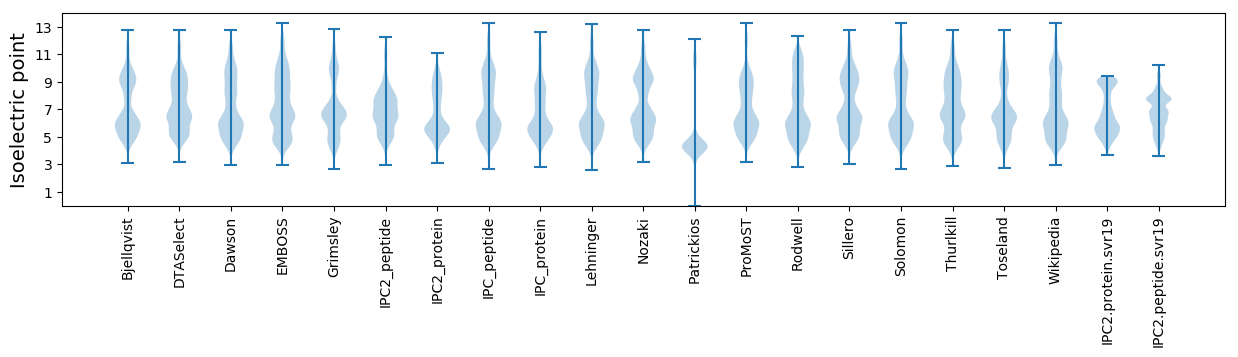
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C3PNC4|A0A0C3PNC4_PHLGI Uncharacterized protein OS=Phlebiopsis gigantea 11061_1 CR5-6 OX=745531 GN=PHLGIDRAFT_104544 PE=3 SV=1
VV1 pKa = 7.11ASCGSHH7 pKa = 4.89RR8 pKa = 11.84TSDD11 pKa = 3.7RR12 pKa = 11.84LCGGGEE18 pKa = 3.75PSVRR22 pKa = 11.84PRR24 pKa = 11.84VLGRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84PRR32 pKa = 11.84HH33 pKa = 5.31PPGLRR38 pKa = 11.84PRR40 pKa = 11.84LPRR43 pKa = 11.84LAPALPPPSLQSRR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84AVRR61 pKa = 11.84PRR63 pKa = 11.84RR64 pKa = 11.84CLHH67 pKa = 5.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84PARR73 pKa = 11.84PAPPPRR79 pKa = 11.84VPGPVRR85 pKa = 11.84PRR87 pKa = 11.84PRR89 pKa = 11.84RR90 pKa = 11.84SLRR93 pKa = 11.84PARR96 pKa = 11.84GPQAAPRR103 pKa = 11.84PDD105 pKa = 3.16AAARR109 pKa = 11.84RR110 pKa = 11.84AARR113 pKa = 11.84PAPSAALRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84PLPAAPARR131 pKa = 11.84PGRR134 pKa = 11.84AHH136 pKa = 6.77AALRR140 pKa = 11.84PRR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84ALPRR148 pKa = 11.84QPRR151 pKa = 11.84LLLSARR157 pKa = 11.84LPPHH161 pKa = 6.86RR162 pKa = 11.84RR163 pKa = 11.84RR164 pKa = 11.84PPQPQAARR172 pKa = 11.84RR173 pKa = 11.84EE174 pKa = 4.19LARR177 pKa = 11.84LGHH180 pKa = 6.17RR181 pKa = 11.84PARR184 pKa = 11.84SSAARR189 pKa = 11.84AVPQVRR195 pKa = 11.84RR196 pKa = 11.84PPRR199 pKa = 11.84APLAGRR205 pKa = 11.84DD206 pKa = 3.46GALDD210 pKa = 3.62DD211 pKa = 4.5RR212 pKa = 11.84QPLGLPRR219 pKa = 11.84QLARR223 pKa = 11.84TLWHH227 pKa = 6.46RR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84SRR232 pKa = 11.84GARR235 pKa = 11.84PAPHH239 pKa = 6.18ARR241 pKa = 11.84ARR243 pKa = 11.84RR244 pKa = 11.84RR245 pKa = 11.84DD246 pKa = 3.05ARR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84PARR255 pKa = 11.84HH256 pKa = 5.69GKK258 pKa = 9.21AAPARR263 pKa = 11.84SRR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84TRR269 pKa = 11.84PGRR272 pKa = 11.84RR273 pKa = 11.84PRR275 pKa = 11.84RR276 pKa = 11.84AVRR279 pKa = 11.84AAARR283 pKa = 11.84IAGPHH288 pKa = 6.65APRR291 pKa = 11.84ALPPAHH297 pKa = 5.78TRR299 pKa = 11.84VLRR302 pKa = 11.84RR303 pKa = 11.84APRR306 pKa = 11.84RR307 pKa = 11.84PPLGPLGAPRR317 pKa = 11.84PALPRR322 pKa = 11.84ARR324 pKa = 11.84HH325 pKa = 5.26PPRR328 pKa = 11.84RR329 pKa = 11.84PVPAALRR336 pKa = 11.84AAALPALPDD345 pKa = 3.37GRR347 pKa = 11.84RR348 pKa = 11.84LARR351 pKa = 11.84ARR353 pKa = 11.84RR354 pKa = 11.84RR355 pKa = 11.84AAVRR359 pKa = 11.84RR360 pKa = 11.84AQAARR365 pKa = 11.84KK366 pKa = 8.86GRR368 pKa = 11.84LRR370 pKa = 11.84RR371 pKa = 11.84VRR373 pKa = 11.84PRR375 pKa = 11.84DD376 pKa = 3.2ARR378 pKa = 11.84RR379 pKa = 11.84AARR382 pKa = 11.84AGLGRR387 pKa = 11.84RR388 pKa = 11.84RR389 pKa = 11.84RR390 pKa = 11.84GLARR394 pKa = 11.84GVGRR398 pKa = 11.84GGARR402 pKa = 11.84GAAWDD407 pKa = 3.64ARR409 pKa = 11.84QGRR412 pKa = 11.84GARR415 pKa = 11.84VEE417 pKa = 3.78RR418 pKa = 11.84GGRR421 pKa = 11.84GRR423 pKa = 11.84PRR425 pKa = 11.84FVSRR429 pKa = 11.84PPTRR433 pKa = 11.84RR434 pKa = 11.84NTCAFQSLLRR444 pKa = 11.84AHH446 pKa = 6.98LEE448 pKa = 4.02VFQGISAHH456 pKa = 5.17VRR458 pKa = 11.84VGYY461 pKa = 10.42
VV1 pKa = 7.11ASCGSHH7 pKa = 4.89RR8 pKa = 11.84TSDD11 pKa = 3.7RR12 pKa = 11.84LCGGGEE18 pKa = 3.75PSVRR22 pKa = 11.84PRR24 pKa = 11.84VLGRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84PRR32 pKa = 11.84HH33 pKa = 5.31PPGLRR38 pKa = 11.84PRR40 pKa = 11.84LPRR43 pKa = 11.84LAPALPPPSLQSRR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84AVRR61 pKa = 11.84PRR63 pKa = 11.84RR64 pKa = 11.84CLHH67 pKa = 5.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84PARR73 pKa = 11.84PAPPPRR79 pKa = 11.84VPGPVRR85 pKa = 11.84PRR87 pKa = 11.84PRR89 pKa = 11.84RR90 pKa = 11.84SLRR93 pKa = 11.84PARR96 pKa = 11.84GPQAAPRR103 pKa = 11.84PDD105 pKa = 3.16AAARR109 pKa = 11.84RR110 pKa = 11.84AARR113 pKa = 11.84PAPSAALRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84PLPAAPARR131 pKa = 11.84PGRR134 pKa = 11.84AHH136 pKa = 6.77AALRR140 pKa = 11.84PRR142 pKa = 11.84RR143 pKa = 11.84RR144 pKa = 11.84ALPRR148 pKa = 11.84QPRR151 pKa = 11.84LLLSARR157 pKa = 11.84LPPHH161 pKa = 6.86RR162 pKa = 11.84RR163 pKa = 11.84RR164 pKa = 11.84PPQPQAARR172 pKa = 11.84RR173 pKa = 11.84EE174 pKa = 4.19LARR177 pKa = 11.84LGHH180 pKa = 6.17RR181 pKa = 11.84PARR184 pKa = 11.84SSAARR189 pKa = 11.84AVPQVRR195 pKa = 11.84RR196 pKa = 11.84PPRR199 pKa = 11.84APLAGRR205 pKa = 11.84DD206 pKa = 3.46GALDD210 pKa = 3.62DD211 pKa = 4.5RR212 pKa = 11.84QPLGLPRR219 pKa = 11.84QLARR223 pKa = 11.84TLWHH227 pKa = 6.46RR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84SRR232 pKa = 11.84GARR235 pKa = 11.84PAPHH239 pKa = 6.18ARR241 pKa = 11.84ARR243 pKa = 11.84RR244 pKa = 11.84RR245 pKa = 11.84DD246 pKa = 3.05ARR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84PARR255 pKa = 11.84HH256 pKa = 5.69GKK258 pKa = 9.21AAPARR263 pKa = 11.84SRR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84TRR269 pKa = 11.84PGRR272 pKa = 11.84RR273 pKa = 11.84PRR275 pKa = 11.84RR276 pKa = 11.84AVRR279 pKa = 11.84AAARR283 pKa = 11.84IAGPHH288 pKa = 6.65APRR291 pKa = 11.84ALPPAHH297 pKa = 5.78TRR299 pKa = 11.84VLRR302 pKa = 11.84RR303 pKa = 11.84APRR306 pKa = 11.84RR307 pKa = 11.84PPLGPLGAPRR317 pKa = 11.84PALPRR322 pKa = 11.84ARR324 pKa = 11.84HH325 pKa = 5.26PPRR328 pKa = 11.84RR329 pKa = 11.84PVPAALRR336 pKa = 11.84AAALPALPDD345 pKa = 3.37GRR347 pKa = 11.84RR348 pKa = 11.84LARR351 pKa = 11.84ARR353 pKa = 11.84RR354 pKa = 11.84RR355 pKa = 11.84AAVRR359 pKa = 11.84RR360 pKa = 11.84AQAARR365 pKa = 11.84KK366 pKa = 8.86GRR368 pKa = 11.84LRR370 pKa = 11.84RR371 pKa = 11.84VRR373 pKa = 11.84PRR375 pKa = 11.84DD376 pKa = 3.2ARR378 pKa = 11.84RR379 pKa = 11.84AARR382 pKa = 11.84AGLGRR387 pKa = 11.84RR388 pKa = 11.84RR389 pKa = 11.84RR390 pKa = 11.84GLARR394 pKa = 11.84GVGRR398 pKa = 11.84GGARR402 pKa = 11.84GAAWDD407 pKa = 3.64ARR409 pKa = 11.84QGRR412 pKa = 11.84GARR415 pKa = 11.84VEE417 pKa = 3.78RR418 pKa = 11.84GGRR421 pKa = 11.84GRR423 pKa = 11.84PRR425 pKa = 11.84FVSRR429 pKa = 11.84PPTRR433 pKa = 11.84RR434 pKa = 11.84NTCAFQSLLRR444 pKa = 11.84AHH446 pKa = 6.98LEE448 pKa = 4.02VFQGISAHH456 pKa = 5.17VRR458 pKa = 11.84VGYY461 pKa = 10.42
Molecular weight: 51.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4868893 |
36 |
4819 |
410.9 |
45.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.365 ± 0.02 | 1.296 ± 0.01 |
5.544 ± 0.018 | 5.749 ± 0.027 |
3.649 ± 0.015 | 6.532 ± 0.023 |
2.604 ± 0.01 | 4.525 ± 0.017 |
4.195 ± 0.021 | 9.286 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.051 ± 0.008 | 3.098 ± 0.011 |
6.604 ± 0.029 | 3.691 ± 0.015 |
6.495 ± 0.025 | 8.388 ± 0.027 |
6.111 ± 0.014 | 6.659 ± 0.016 |
1.47 ± 0.009 | 2.681 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
