
Tistrella mobilis (strain KA081020-065)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Tistrella; Tistrella mobilis
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
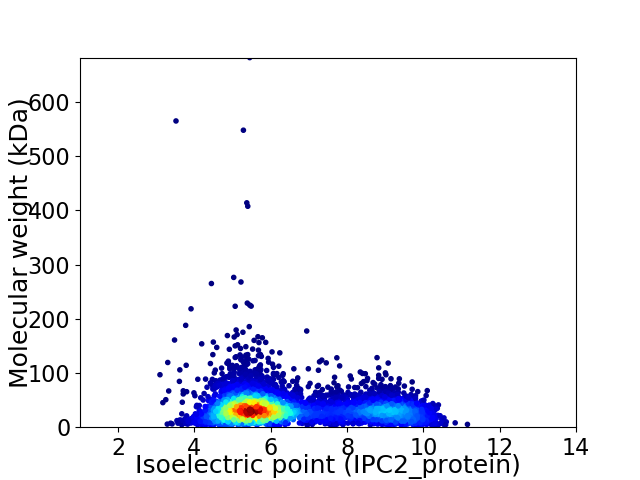
Virtual 2D-PAGE plot for 5778 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
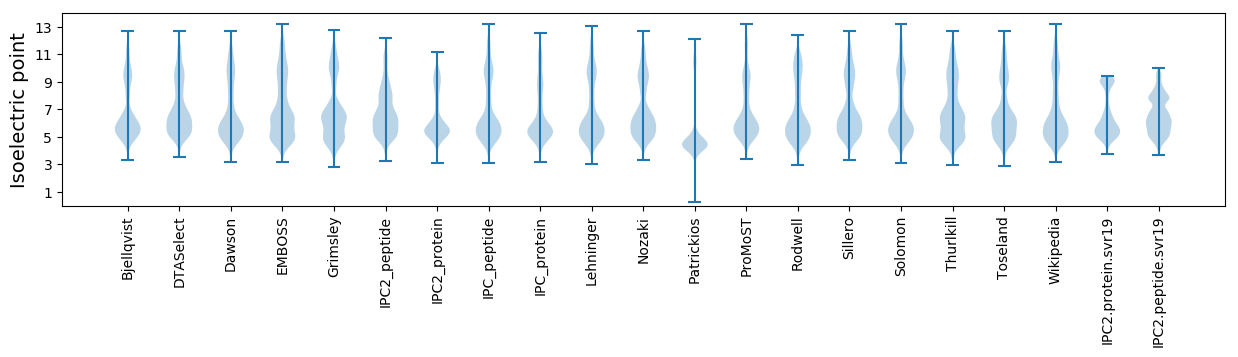
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3TV30|I3TV30_TISMK Copper resistance protein CopC OS=Tistrella mobilis (strain KA081020-065) OX=1110502 GN=copC PE=3 SV=1
MM1 pKa = 8.05ADD3 pKa = 3.26TPYY6 pKa = 10.56ATLTPPSGWSVYY18 pKa = 10.5SNQGAEE24 pKa = 4.35SGLKK28 pKa = 7.45TTVSLSSGEE37 pKa = 4.1IVGGDD42 pKa = 3.59PAFLDD47 pKa = 4.33SIADD51 pKa = 3.45AFYY54 pKa = 10.78EE55 pKa = 4.35VALTAVEE62 pKa = 4.93AYY64 pKa = 7.2ITPSNIEE71 pKa = 4.66DD72 pKa = 4.11PLQNWGDD79 pKa = 3.77FLTSLKK85 pKa = 10.69SNTVIRR91 pKa = 11.84DD92 pKa = 3.61ALDD95 pKa = 3.85SIANVTAVQAVQAVAGNTVQTSGSALGEE123 pKa = 4.18LQDD126 pKa = 4.46EE127 pKa = 4.65TLQDD131 pKa = 3.19ATDD134 pKa = 4.76AIFSEE139 pKa = 4.61LNSGNVEE146 pKa = 4.1PGSLLGWYY154 pKa = 9.54QYY156 pKa = 10.25TIHH159 pKa = 7.41TITQLLTSTTLQFQKK174 pKa = 10.79SVVGNAVEE182 pKa = 4.64IYY184 pKa = 10.55LGLDD188 pKa = 3.31GQMVAASNSSSITYY202 pKa = 9.97SGNNSVVVQGGAADD216 pKa = 4.49DD217 pKa = 4.57LLTSGSRR224 pKa = 11.84DD225 pKa = 3.43DD226 pKa = 5.52VLVGWAGNDD235 pKa = 2.77IMIASEE241 pKa = 4.58GKK243 pKa = 8.4DD244 pKa = 3.27TYY246 pKa = 10.96IGGDD250 pKa = 3.3GTDD253 pKa = 3.2TVDD256 pKa = 2.95YY257 pKa = 11.29SGFHH261 pKa = 6.73GFATPLKK268 pKa = 10.37ASLATGKK275 pKa = 8.78GTSGPATGHH284 pKa = 5.98SYY286 pKa = 11.25VGVEE290 pKa = 4.0VLIGTAGGDD299 pKa = 3.47DD300 pKa = 3.79LTANGAGTTLYY311 pKa = 10.91GGGGNDD317 pKa = 3.53VLRR320 pKa = 11.84DD321 pKa = 3.57TSTTGSATLNGGDD334 pKa = 4.13GNDD337 pKa = 3.34TASYY341 pKa = 10.16FGRR344 pKa = 11.84TQGITASLSAPGDD357 pKa = 3.49TTGYY361 pKa = 10.22LISIEE366 pKa = 4.03NLYY369 pKa = 8.57GTDD372 pKa = 3.71YY373 pKa = 11.69NDD375 pKa = 3.31VLVGNSEE382 pKa = 4.06NNILTGLEE390 pKa = 4.26GNDD393 pKa = 3.42TLVGRR398 pKa = 11.84GGADD402 pKa = 3.07TLDD405 pKa = 3.54GRR407 pKa = 11.84GGSDD411 pKa = 2.9TASYY415 pKa = 10.82QSASSSVTADD425 pKa = 3.13LRR427 pKa = 11.84AGNGTQGDD435 pKa = 3.97AAGDD439 pKa = 3.64TLISIEE445 pKa = 4.26NLTGSAYY452 pKa = 10.39GDD454 pKa = 3.62KK455 pKa = 10.89LYY457 pKa = 11.12GDD459 pKa = 3.9SAANVLRR466 pKa = 11.84GGNGDD471 pKa = 3.43DD472 pKa = 3.63ALRR475 pKa = 11.84GRR477 pKa = 11.84GGADD481 pKa = 2.94TLDD484 pKa = 3.93GGNGFDD490 pKa = 3.48FASYY494 pKa = 11.13SDD496 pKa = 3.39ATASVTVDD504 pKa = 3.77LASGKK509 pKa = 10.21GLAGEE514 pKa = 4.31ALGDD518 pKa = 3.64TLVSIEE524 pKa = 4.01GLAGSGYY531 pKa = 10.8DD532 pKa = 5.63DD533 pKa = 5.58LLQGDD538 pKa = 4.15GNDD541 pKa = 3.05NYY543 pKa = 10.88FYY545 pKa = 11.44GNGGNDD551 pKa = 3.14ILRR554 pKa = 11.84GRR556 pKa = 11.84GGADD560 pKa = 2.91TFDD563 pKa = 4.2GGSGLDD569 pKa = 3.36FATYY573 pKa = 10.56SDD575 pKa = 3.85ASASVVADD583 pKa = 4.35LASGGISGEE592 pKa = 4.18AEE594 pKa = 3.54GDD596 pKa = 3.73VYY598 pKa = 11.07ISIEE602 pKa = 4.03GLTGTSYY609 pKa = 12.08ADD611 pKa = 3.77TLHH614 pKa = 7.07GDD616 pKa = 3.33AGSNTLVGRR625 pKa = 11.84DD626 pKa = 3.67GDD628 pKa = 4.62DD629 pKa = 2.99ILNGRR634 pKa = 11.84RR635 pKa = 11.84GADD638 pKa = 3.22QLIGGDD644 pKa = 3.54GFDD647 pKa = 3.26FATYY651 pKa = 10.31SEE653 pKa = 4.45ASAAVAVDD661 pKa = 4.22LASQRR666 pKa = 11.84GTAGEE671 pKa = 4.17AKK673 pKa = 10.36DD674 pKa = 5.44DD675 pKa = 3.63ILTEE679 pKa = 3.94IEE681 pKa = 4.13GLIGSAYY688 pKa = 10.47GDD690 pKa = 3.37TLSGNKK696 pKa = 8.69EE697 pKa = 4.01VNHH700 pKa = 6.25LRR702 pKa = 11.84GEE704 pKa = 4.28AGNDD708 pKa = 3.3VLDD711 pKa = 4.15GRR713 pKa = 11.84GGADD717 pKa = 3.79LLQGGAGADD726 pKa = 3.47QLTGGTGADD735 pKa = 2.96NFLYY739 pKa = 10.97LNLSDD744 pKa = 3.96SGKK747 pKa = 9.0TSGTWDD753 pKa = 3.82TITDD757 pKa = 4.35FSQTDD762 pKa = 3.28GDD764 pKa = 4.58KK765 pKa = 10.62IDD767 pKa = 3.72VSAIDD772 pKa = 4.63AYY774 pKa = 11.18VGSPGNQTFTFIGSAGYY791 pKa = 10.3SGTPGEE797 pKa = 4.24LRR799 pKa = 11.84ANVSNGYY806 pKa = 7.79TYY808 pKa = 10.55IGVDD812 pKa = 3.13VDD814 pKa = 4.06GDD816 pKa = 3.72QASDD820 pKa = 3.24MVIRR824 pKa = 11.84LNGIHH829 pKa = 6.86TLTAADD835 pKa = 5.33FILL838 pKa = 5.1
MM1 pKa = 8.05ADD3 pKa = 3.26TPYY6 pKa = 10.56ATLTPPSGWSVYY18 pKa = 10.5SNQGAEE24 pKa = 4.35SGLKK28 pKa = 7.45TTVSLSSGEE37 pKa = 4.1IVGGDD42 pKa = 3.59PAFLDD47 pKa = 4.33SIADD51 pKa = 3.45AFYY54 pKa = 10.78EE55 pKa = 4.35VALTAVEE62 pKa = 4.93AYY64 pKa = 7.2ITPSNIEE71 pKa = 4.66DD72 pKa = 4.11PLQNWGDD79 pKa = 3.77FLTSLKK85 pKa = 10.69SNTVIRR91 pKa = 11.84DD92 pKa = 3.61ALDD95 pKa = 3.85SIANVTAVQAVQAVAGNTVQTSGSALGEE123 pKa = 4.18LQDD126 pKa = 4.46EE127 pKa = 4.65TLQDD131 pKa = 3.19ATDD134 pKa = 4.76AIFSEE139 pKa = 4.61LNSGNVEE146 pKa = 4.1PGSLLGWYY154 pKa = 9.54QYY156 pKa = 10.25TIHH159 pKa = 7.41TITQLLTSTTLQFQKK174 pKa = 10.79SVVGNAVEE182 pKa = 4.64IYY184 pKa = 10.55LGLDD188 pKa = 3.31GQMVAASNSSSITYY202 pKa = 9.97SGNNSVVVQGGAADD216 pKa = 4.49DD217 pKa = 4.57LLTSGSRR224 pKa = 11.84DD225 pKa = 3.43DD226 pKa = 5.52VLVGWAGNDD235 pKa = 2.77IMIASEE241 pKa = 4.58GKK243 pKa = 8.4DD244 pKa = 3.27TYY246 pKa = 10.96IGGDD250 pKa = 3.3GTDD253 pKa = 3.2TVDD256 pKa = 2.95YY257 pKa = 11.29SGFHH261 pKa = 6.73GFATPLKK268 pKa = 10.37ASLATGKK275 pKa = 8.78GTSGPATGHH284 pKa = 5.98SYY286 pKa = 11.25VGVEE290 pKa = 4.0VLIGTAGGDD299 pKa = 3.47DD300 pKa = 3.79LTANGAGTTLYY311 pKa = 10.91GGGGNDD317 pKa = 3.53VLRR320 pKa = 11.84DD321 pKa = 3.57TSTTGSATLNGGDD334 pKa = 4.13GNDD337 pKa = 3.34TASYY341 pKa = 10.16FGRR344 pKa = 11.84TQGITASLSAPGDD357 pKa = 3.49TTGYY361 pKa = 10.22LISIEE366 pKa = 4.03NLYY369 pKa = 8.57GTDD372 pKa = 3.71YY373 pKa = 11.69NDD375 pKa = 3.31VLVGNSEE382 pKa = 4.06NNILTGLEE390 pKa = 4.26GNDD393 pKa = 3.42TLVGRR398 pKa = 11.84GGADD402 pKa = 3.07TLDD405 pKa = 3.54GRR407 pKa = 11.84GGSDD411 pKa = 2.9TASYY415 pKa = 10.82QSASSSVTADD425 pKa = 3.13LRR427 pKa = 11.84AGNGTQGDD435 pKa = 3.97AAGDD439 pKa = 3.64TLISIEE445 pKa = 4.26NLTGSAYY452 pKa = 10.39GDD454 pKa = 3.62KK455 pKa = 10.89LYY457 pKa = 11.12GDD459 pKa = 3.9SAANVLRR466 pKa = 11.84GGNGDD471 pKa = 3.43DD472 pKa = 3.63ALRR475 pKa = 11.84GRR477 pKa = 11.84GGADD481 pKa = 2.94TLDD484 pKa = 3.93GGNGFDD490 pKa = 3.48FASYY494 pKa = 11.13SDD496 pKa = 3.39ATASVTVDD504 pKa = 3.77LASGKK509 pKa = 10.21GLAGEE514 pKa = 4.31ALGDD518 pKa = 3.64TLVSIEE524 pKa = 4.01GLAGSGYY531 pKa = 10.8DD532 pKa = 5.63DD533 pKa = 5.58LLQGDD538 pKa = 4.15GNDD541 pKa = 3.05NYY543 pKa = 10.88FYY545 pKa = 11.44GNGGNDD551 pKa = 3.14ILRR554 pKa = 11.84GRR556 pKa = 11.84GGADD560 pKa = 2.91TFDD563 pKa = 4.2GGSGLDD569 pKa = 3.36FATYY573 pKa = 10.56SDD575 pKa = 3.85ASASVVADD583 pKa = 4.35LASGGISGEE592 pKa = 4.18AEE594 pKa = 3.54GDD596 pKa = 3.73VYY598 pKa = 11.07ISIEE602 pKa = 4.03GLTGTSYY609 pKa = 12.08ADD611 pKa = 3.77TLHH614 pKa = 7.07GDD616 pKa = 3.33AGSNTLVGRR625 pKa = 11.84DD626 pKa = 3.67GDD628 pKa = 4.62DD629 pKa = 2.99ILNGRR634 pKa = 11.84RR635 pKa = 11.84GADD638 pKa = 3.22QLIGGDD644 pKa = 3.54GFDD647 pKa = 3.26FATYY651 pKa = 10.31SEE653 pKa = 4.45ASAAVAVDD661 pKa = 4.22LASQRR666 pKa = 11.84GTAGEE671 pKa = 4.17AKK673 pKa = 10.36DD674 pKa = 5.44DD675 pKa = 3.63ILTEE679 pKa = 3.94IEE681 pKa = 4.13GLIGSAYY688 pKa = 10.47GDD690 pKa = 3.37TLSGNKK696 pKa = 8.69EE697 pKa = 4.01VNHH700 pKa = 6.25LRR702 pKa = 11.84GEE704 pKa = 4.28AGNDD708 pKa = 3.3VLDD711 pKa = 4.15GRR713 pKa = 11.84GGADD717 pKa = 3.79LLQGGAGADD726 pKa = 3.47QLTGGTGADD735 pKa = 2.96NFLYY739 pKa = 10.97LNLSDD744 pKa = 3.96SGKK747 pKa = 9.0TSGTWDD753 pKa = 3.82TITDD757 pKa = 4.35FSQTDD762 pKa = 3.28GDD764 pKa = 4.58KK765 pKa = 10.62IDD767 pKa = 3.72VSAIDD772 pKa = 4.63AYY774 pKa = 11.18VGSPGNQTFTFIGSAGYY791 pKa = 10.3SGTPGEE797 pKa = 4.24LRR799 pKa = 11.84ANVSNGYY806 pKa = 7.79TYY808 pKa = 10.55IGVDD812 pKa = 3.13VDD814 pKa = 4.06GDD816 pKa = 3.72QASDD820 pKa = 3.24MVIRR824 pKa = 11.84LNGIHH829 pKa = 6.86TLTAADD835 pKa = 5.33FILL838 pKa = 5.1
Molecular weight: 84.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3TPA1|I3TPA1_TISMK Uncharacterized protein OS=Tistrella mobilis (strain KA081020-065) OX=1110502 GN=TMO_2751 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.06VLANRR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.9RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 8.95RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.63GFRR19 pKa = 11.84SRR21 pKa = 11.84MATVGGRR28 pKa = 11.84KK29 pKa = 9.06VLANRR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.9RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1925682 |
37 |
6469 |
333.3 |
35.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.444 ± 0.066 | 0.725 ± 0.01 |
6.164 ± 0.034 | 5.191 ± 0.027 |
3.245 ± 0.021 | 9.318 ± 0.033 |
2.005 ± 0.016 | 4.826 ± 0.02 |
2.031 ± 0.028 | 10.57 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.488 ± 0.017 | 1.871 ± 0.016 |
5.744 ± 0.031 | 2.63 ± 0.019 |
8.17 ± 0.04 | 4.295 ± 0.023 |
5.391 ± 0.032 | 7.629 ± 0.033 |
1.344 ± 0.014 | 1.919 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
