
Psilocybe cyanescens
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Strophariaceae; Psilocybe
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
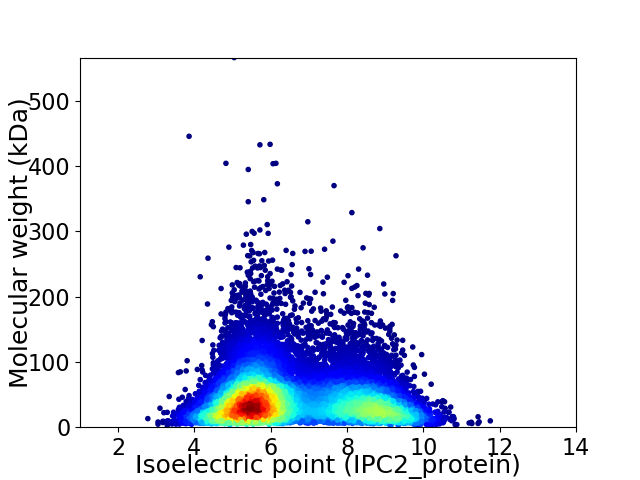
Virtual 2D-PAGE plot for 15931 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A409XBD9|A0A409XBD9_PSICY GMC_OxRdtase_N domain-containing protein OS=Psilocybe cyanescens OX=93625 GN=CVT25_000843 PE=3 SV=1
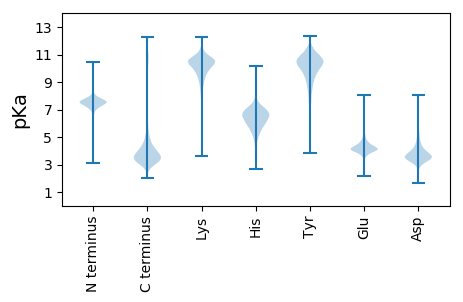
MM1 pKa = 6.99TWRR4 pKa = 11.84TAEE7 pKa = 4.16TLAHH11 pKa = 6.86DD12 pKa = 4.16CQFAHH17 pKa = 6.4SASVLIDD24 pKa = 3.41GSFLGPITHH33 pKa = 6.6HH34 pKa = 5.57TAISIDD40 pKa = 3.64NQLALTLGFVSTAAAWNSLSDD61 pKa = 4.03EE62 pKa = 4.82DD63 pKa = 5.11EE64 pKa = 4.93DD65 pKa = 6.33DD66 pKa = 3.99NFIPADD72 pKa = 3.7ALPTNKK78 pKa = 10.13ASTNPPLSIAAQVAEE93 pKa = 4.12LWLRR97 pKa = 11.84DD98 pKa = 3.55SSPEE102 pKa = 3.79VSSPSPQHH110 pKa = 6.19NFHH113 pKa = 6.78TGPNDD118 pKa = 3.82LAVAWAISSDD128 pKa = 3.54EE129 pKa = 4.46DD130 pKa = 3.84LASDD134 pKa = 4.32GAYY137 pKa = 8.74PNSVISIEE145 pKa = 4.28AVCHH149 pKa = 5.17VLSHH153 pKa = 6.57EE154 pKa = 4.28NLNILASVGATDD166 pKa = 5.88AGAVWLNSLDD176 pKa = 3.76SGANNSLGPAAEE188 pKa = 4.16QQYY191 pKa = 8.26STIFPNPITPPVGLAEE207 pKa = 4.3LTVVWYY213 pKa = 10.83SSDD216 pKa = 3.6KK217 pKa = 11.05DD218 pKa = 3.54QSSYY222 pKa = 11.03SASAPPSDD230 pKa = 3.12WHH232 pKa = 6.93LSQADD237 pKa = 3.45SSMRR241 pKa = 11.84PAQVYY246 pKa = 7.66MAVDD250 pKa = 3.92SVTPSHH256 pKa = 6.38ITLPCPFTPTFFDD269 pKa = 4.48FVDD272 pKa = 4.2DD273 pKa = 3.45TWLPAEE279 pKa = 4.7EE280 pKa = 5.49DD281 pKa = 3.55SGDD284 pKa = 3.9SNNEE288 pKa = 2.92QWAADD293 pKa = 3.67TDD295 pKa = 3.94DD296 pKa = 3.73TEE298 pKa = 4.74EE299 pKa = 4.04FVLL302 pKa = 5.13
MM1 pKa = 6.99TWRR4 pKa = 11.84TAEE7 pKa = 4.16TLAHH11 pKa = 6.86DD12 pKa = 4.16CQFAHH17 pKa = 6.4SASVLIDD24 pKa = 3.41GSFLGPITHH33 pKa = 6.6HH34 pKa = 5.57TAISIDD40 pKa = 3.64NQLALTLGFVSTAAAWNSLSDD61 pKa = 4.03EE62 pKa = 4.82DD63 pKa = 5.11EE64 pKa = 4.93DD65 pKa = 6.33DD66 pKa = 3.99NFIPADD72 pKa = 3.7ALPTNKK78 pKa = 10.13ASTNPPLSIAAQVAEE93 pKa = 4.12LWLRR97 pKa = 11.84DD98 pKa = 3.55SSPEE102 pKa = 3.79VSSPSPQHH110 pKa = 6.19NFHH113 pKa = 6.78TGPNDD118 pKa = 3.82LAVAWAISSDD128 pKa = 3.54EE129 pKa = 4.46DD130 pKa = 3.84LASDD134 pKa = 4.32GAYY137 pKa = 8.74PNSVISIEE145 pKa = 4.28AVCHH149 pKa = 5.17VLSHH153 pKa = 6.57EE154 pKa = 4.28NLNILASVGATDD166 pKa = 5.88AGAVWLNSLDD176 pKa = 3.76SGANNSLGPAAEE188 pKa = 4.16QQYY191 pKa = 8.26STIFPNPITPPVGLAEE207 pKa = 4.3LTVVWYY213 pKa = 10.83SSDD216 pKa = 3.6KK217 pKa = 11.05DD218 pKa = 3.54QSSYY222 pKa = 11.03SASAPPSDD230 pKa = 3.12WHH232 pKa = 6.93LSQADD237 pKa = 3.45SSMRR241 pKa = 11.84PAQVYY246 pKa = 7.66MAVDD250 pKa = 3.92SVTPSHH256 pKa = 6.38ITLPCPFTPTFFDD269 pKa = 4.48FVDD272 pKa = 4.2DD273 pKa = 3.45TWLPAEE279 pKa = 4.7EE280 pKa = 5.49DD281 pKa = 3.55SGDD284 pKa = 3.9SNNEE288 pKa = 2.92QWAADD293 pKa = 3.67TDD295 pKa = 3.94DD296 pKa = 3.73TEE298 pKa = 4.74EE299 pKa = 4.04FVLL302 pKa = 5.13
Molecular weight: 32.29 kDa
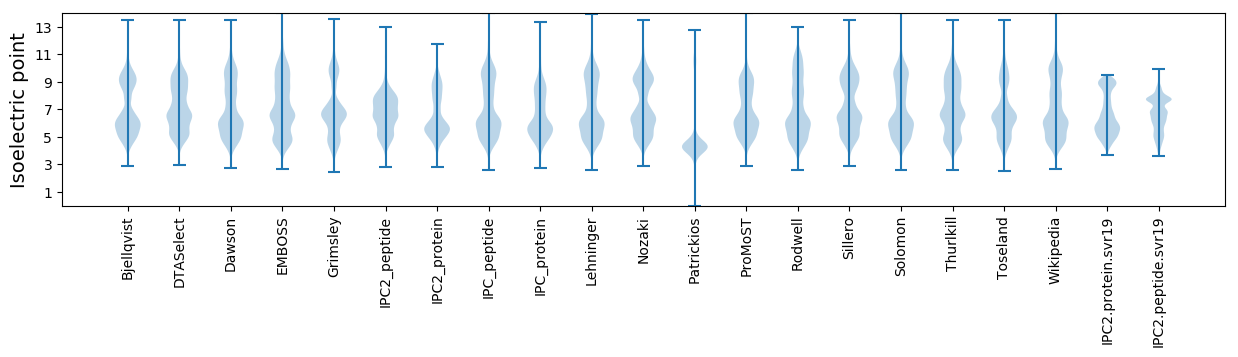
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A409WEU8|A0A409WEU8_PSICY Uncharacterized protein OS=Psilocybe cyanescens OX=93625 GN=CVT25_014831 PE=4 SV=1
MM1 pKa = 7.44RR2 pKa = 11.84QRR4 pKa = 11.84TRR6 pKa = 11.84MTAMATPMRR15 pKa = 11.84TTAMTRR21 pKa = 11.84SWTRR25 pKa = 11.84MTATATTRR33 pKa = 11.84TRR35 pKa = 11.84TRR37 pKa = 11.84TTTRR41 pKa = 11.84TARR44 pKa = 11.84ARR46 pKa = 11.84TTAA49 pKa = 3.37
MM1 pKa = 7.44RR2 pKa = 11.84QRR4 pKa = 11.84TRR6 pKa = 11.84MTAMATPMRR15 pKa = 11.84TTAMTRR21 pKa = 11.84SWTRR25 pKa = 11.84MTATATTRR33 pKa = 11.84TRR35 pKa = 11.84TRR37 pKa = 11.84TTTRR41 pKa = 11.84TARR44 pKa = 11.84ARR46 pKa = 11.84TTAA49 pKa = 3.37
Molecular weight: 5.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6841066 |
16 |
5056 |
429.4 |
47.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.129 ± 0.019 | 1.211 ± 0.007 |
5.557 ± 0.015 | 5.682 ± 0.02 |
3.818 ± 0.013 | 6.354 ± 0.019 |
2.607 ± 0.01 | 5.307 ± 0.017 |
4.71 ± 0.019 | 9.128 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.081 ± 0.007 | 3.89 ± 0.011 |
6.354 ± 0.025 | 3.822 ± 0.014 |
5.755 ± 0.017 | 9.111 ± 0.02 |
6.221 ± 0.013 | 6.185 ± 0.015 |
1.358 ± 0.006 | 2.713 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
