
Rhizoctonia solani virus 717
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Betapartitivirus
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
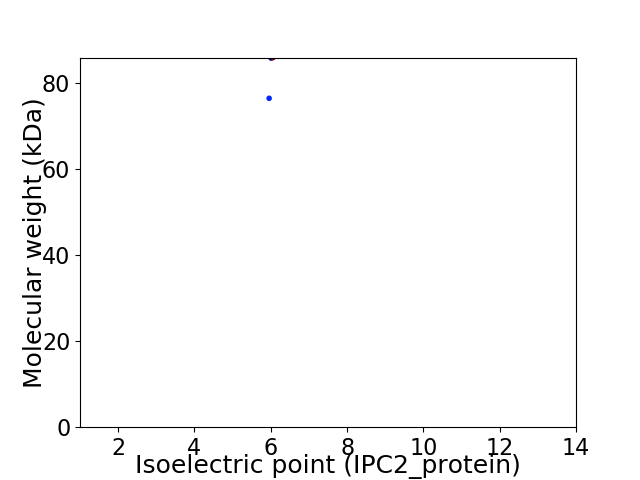
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9Q9X2|Q9Q9X2_9VIRU Putative RNA-dependent RNA polymerase OS=Rhizoctonia solani virus 717 OX=46618 PE=4 SV=1
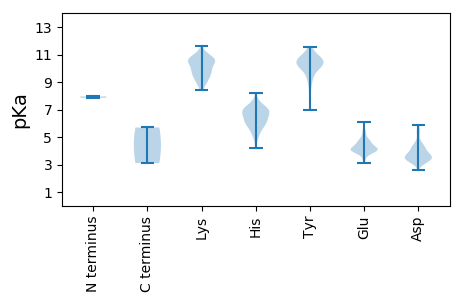
MM1 pKa = 7.85LYY3 pKa = 10.41NFKK6 pKa = 11.1AFISKK11 pKa = 9.74VSHH14 pKa = 6.96SIRR17 pKa = 11.84DD18 pKa = 3.98YY19 pKa = 10.39LTEE22 pKa = 4.18AYY24 pKa = 9.2IRR26 pKa = 11.84RR27 pKa = 11.84NFEE30 pKa = 3.34WSNFNKK36 pKa = 10.28LHH38 pKa = 6.65TDD40 pKa = 3.53PSTAYY45 pKa = 10.7YY46 pKa = 10.29DD47 pKa = 3.27IRR49 pKa = 11.84DD50 pKa = 3.82PSNVRR55 pKa = 11.84AYY57 pKa = 10.19HH58 pKa = 6.18ALFGTRR64 pKa = 11.84RR65 pKa = 11.84QPPPKK70 pKa = 9.94QIEE73 pKa = 4.35LEE75 pKa = 4.2SYY77 pKa = 9.18MLNLIHH83 pKa = 7.49RR84 pKa = 11.84NAIKK88 pKa = 10.32NRR90 pKa = 11.84PFQFYY95 pKa = 10.26TDD97 pKa = 4.54LFTDD101 pKa = 3.51TDD103 pKa = 4.42LPTDD107 pKa = 5.33RR108 pKa = 11.84IPKK111 pKa = 9.72PGIEE115 pKa = 4.04LVNAYY120 pKa = 7.22FHH122 pKa = 7.26PGNVIRR128 pKa = 11.84PNPEE132 pKa = 3.13FSKK135 pKa = 10.91QLTPDD140 pKa = 3.26MEE142 pKa = 6.07LDD144 pKa = 3.38QFEE147 pKa = 4.95SYY149 pKa = 10.73IPGDD153 pKa = 3.32IDD155 pKa = 3.73YY156 pKa = 10.79GPPIDD161 pKa = 5.58NEE163 pKa = 4.41LLTLIYY169 pKa = 10.1RR170 pKa = 11.84KK171 pKa = 9.85YY172 pKa = 9.83PQYY175 pKa = 10.98LDD177 pKa = 5.46PITKK181 pKa = 8.61YY182 pKa = 10.31CRR184 pKa = 11.84PAGTTDD190 pKa = 2.59ATFRR194 pKa = 11.84DD195 pKa = 4.49FNKK198 pKa = 10.35EE199 pKa = 3.84QIKK202 pKa = 9.31TEE204 pKa = 4.22DD205 pKa = 3.25SDD207 pKa = 4.58ALRR210 pKa = 11.84LSNIMDD216 pKa = 5.9LIHH219 pKa = 6.88EE220 pKa = 4.76FMNITPYY227 pKa = 10.2MPLHH231 pKa = 6.02FVDD234 pKa = 4.25TFYY237 pKa = 11.5CKK239 pKa = 10.63LPLVTGTGYY248 pKa = 10.78HH249 pKa = 5.8NRR251 pKa = 11.84HH252 pKa = 5.7SYY254 pKa = 9.57FRR256 pKa = 11.84RR257 pKa = 11.84SFAHH261 pKa = 6.2FCHH264 pKa = 6.84PEE266 pKa = 4.07LYY268 pKa = 10.47AEE270 pKa = 4.65KK271 pKa = 8.44PTSKK275 pKa = 10.74GYY277 pKa = 10.29FFNATKK283 pKa = 10.48HH284 pKa = 4.88EE285 pKa = 3.95NRR287 pKa = 11.84FLVHH291 pKa = 6.88KK292 pKa = 9.18IKK294 pKa = 10.8HH295 pKa = 5.41SGYY298 pKa = 9.39PFDD301 pKa = 4.22FTSDD305 pKa = 3.1SDD307 pKa = 4.3RR308 pKa = 11.84NSKK311 pKa = 9.03MADD314 pKa = 3.43FLKK317 pKa = 10.75SFPTMMFTRR326 pKa = 11.84NHH328 pKa = 5.28ISKK331 pKa = 10.37RR332 pKa = 11.84DD333 pKa = 3.44GTLKK337 pKa = 10.16VRR339 pKa = 11.84PVYY342 pKa = 10.59AVDD345 pKa = 4.16EE346 pKa = 4.51LFLDD350 pKa = 4.56LEE352 pKa = 4.57CMLAFPATVQARR364 pKa = 11.84KK365 pKa = 9.47PEE367 pKa = 4.1CCIMYY372 pKa = 10.3GLEE375 pKa = 4.27TIRR378 pKa = 11.84GSNIKK383 pKa = 10.13LDD385 pKa = 3.71SLAQGFISFATIDD398 pKa = 3.19WSGYY402 pKa = 6.98DD403 pKa = 3.9QRR405 pKa = 11.84LPWFIVRR412 pKa = 11.84AFFFIYY418 pKa = 10.26LPSLLIISHH427 pKa = 6.6GYY429 pKa = 8.34MPTSEE434 pKa = 4.69YY435 pKa = 10.64EE436 pKa = 4.06DD437 pKa = 3.28TSMNISDD444 pKa = 3.51MFTRR448 pKa = 11.84FFNLINFTATWYY460 pKa = 10.72FNMVFLSADD469 pKa = 3.08GFAFRR474 pKa = 11.84RR475 pKa = 11.84QFAGVPSGMLLTQFLDD491 pKa = 3.73SFGNLYY497 pKa = 10.67LIIDD501 pKa = 3.87SLLEE505 pKa = 3.99FGCTYY510 pKa = 11.56DD511 pKa = 5.14DD512 pKa = 4.17IKK514 pKa = 11.59SLMLFIMGDD523 pKa = 3.62DD524 pKa = 3.55NSIFTNWTIDD534 pKa = 3.41KK535 pKa = 9.92LHH537 pKa = 6.93DD538 pKa = 4.49FISFMEE544 pKa = 4.63RR545 pKa = 11.84YY546 pKa = 9.0CLKK549 pKa = 9.56RR550 pKa = 11.84WNMHH554 pKa = 5.18LSKK557 pKa = 9.17TKK559 pKa = 10.71SVITTLRR566 pKa = 11.84SKK568 pKa = 10.66IEE570 pKa = 4.02TLSYY574 pKa = 10.17RR575 pKa = 11.84CNFGKK580 pKa = 9.73PRR582 pKa = 11.84RR583 pKa = 11.84DD584 pKa = 3.36VEE586 pKa = 4.11KK587 pKa = 10.91LIAQLVYY594 pKa = 10.03PEE596 pKa = 5.38HH597 pKa = 6.25GLKK600 pKa = 9.66PQFMSSRR607 pKa = 11.84AIGLAYY613 pKa = 10.1ASCAQDD619 pKa = 3.26STFHH623 pKa = 6.11EE624 pKa = 5.44FCHH627 pKa = 6.49DD628 pKa = 3.66VYY630 pKa = 11.2RR631 pKa = 11.84LYY633 pKa = 11.25LPVADD638 pKa = 5.5LSPAAIRR645 pKa = 11.84NTRR648 pKa = 11.84VWILKK653 pKa = 9.15LLEE656 pKa = 4.17MEE658 pKa = 4.2EE659 pKa = 4.36TEE661 pKa = 4.83ALIPLDD667 pKa = 4.11HH668 pKa = 7.15FPTMSEE674 pKa = 3.84IQHH677 pKa = 6.31LLSYY681 pKa = 9.88YY682 pKa = 10.19HH683 pKa = 6.91GPLRR687 pKa = 11.84PEE689 pKa = 4.17PKK691 pKa = 8.81WNYY694 pKa = 10.04AHH696 pKa = 7.2FPQDD700 pKa = 3.06PDD702 pKa = 3.88FRR704 pKa = 11.84PKK706 pKa = 10.84DD707 pKa = 3.77YY708 pKa = 10.59VTLLDD713 pKa = 3.52YY714 pKa = 10.49MEE716 pKa = 5.31RR717 pKa = 11.84NNISFPEE724 pKa = 4.7LINFTVV730 pKa = 3.1
MM1 pKa = 7.85LYY3 pKa = 10.41NFKK6 pKa = 11.1AFISKK11 pKa = 9.74VSHH14 pKa = 6.96SIRR17 pKa = 11.84DD18 pKa = 3.98YY19 pKa = 10.39LTEE22 pKa = 4.18AYY24 pKa = 9.2IRR26 pKa = 11.84RR27 pKa = 11.84NFEE30 pKa = 3.34WSNFNKK36 pKa = 10.28LHH38 pKa = 6.65TDD40 pKa = 3.53PSTAYY45 pKa = 10.7YY46 pKa = 10.29DD47 pKa = 3.27IRR49 pKa = 11.84DD50 pKa = 3.82PSNVRR55 pKa = 11.84AYY57 pKa = 10.19HH58 pKa = 6.18ALFGTRR64 pKa = 11.84RR65 pKa = 11.84QPPPKK70 pKa = 9.94QIEE73 pKa = 4.35LEE75 pKa = 4.2SYY77 pKa = 9.18MLNLIHH83 pKa = 7.49RR84 pKa = 11.84NAIKK88 pKa = 10.32NRR90 pKa = 11.84PFQFYY95 pKa = 10.26TDD97 pKa = 4.54LFTDD101 pKa = 3.51TDD103 pKa = 4.42LPTDD107 pKa = 5.33RR108 pKa = 11.84IPKK111 pKa = 9.72PGIEE115 pKa = 4.04LVNAYY120 pKa = 7.22FHH122 pKa = 7.26PGNVIRR128 pKa = 11.84PNPEE132 pKa = 3.13FSKK135 pKa = 10.91QLTPDD140 pKa = 3.26MEE142 pKa = 6.07LDD144 pKa = 3.38QFEE147 pKa = 4.95SYY149 pKa = 10.73IPGDD153 pKa = 3.32IDD155 pKa = 3.73YY156 pKa = 10.79GPPIDD161 pKa = 5.58NEE163 pKa = 4.41LLTLIYY169 pKa = 10.1RR170 pKa = 11.84KK171 pKa = 9.85YY172 pKa = 9.83PQYY175 pKa = 10.98LDD177 pKa = 5.46PITKK181 pKa = 8.61YY182 pKa = 10.31CRR184 pKa = 11.84PAGTTDD190 pKa = 2.59ATFRR194 pKa = 11.84DD195 pKa = 4.49FNKK198 pKa = 10.35EE199 pKa = 3.84QIKK202 pKa = 9.31TEE204 pKa = 4.22DD205 pKa = 3.25SDD207 pKa = 4.58ALRR210 pKa = 11.84LSNIMDD216 pKa = 5.9LIHH219 pKa = 6.88EE220 pKa = 4.76FMNITPYY227 pKa = 10.2MPLHH231 pKa = 6.02FVDD234 pKa = 4.25TFYY237 pKa = 11.5CKK239 pKa = 10.63LPLVTGTGYY248 pKa = 10.78HH249 pKa = 5.8NRR251 pKa = 11.84HH252 pKa = 5.7SYY254 pKa = 9.57FRR256 pKa = 11.84RR257 pKa = 11.84SFAHH261 pKa = 6.2FCHH264 pKa = 6.84PEE266 pKa = 4.07LYY268 pKa = 10.47AEE270 pKa = 4.65KK271 pKa = 8.44PTSKK275 pKa = 10.74GYY277 pKa = 10.29FFNATKK283 pKa = 10.48HH284 pKa = 4.88EE285 pKa = 3.95NRR287 pKa = 11.84FLVHH291 pKa = 6.88KK292 pKa = 9.18IKK294 pKa = 10.8HH295 pKa = 5.41SGYY298 pKa = 9.39PFDD301 pKa = 4.22FTSDD305 pKa = 3.1SDD307 pKa = 4.3RR308 pKa = 11.84NSKK311 pKa = 9.03MADD314 pKa = 3.43FLKK317 pKa = 10.75SFPTMMFTRR326 pKa = 11.84NHH328 pKa = 5.28ISKK331 pKa = 10.37RR332 pKa = 11.84DD333 pKa = 3.44GTLKK337 pKa = 10.16VRR339 pKa = 11.84PVYY342 pKa = 10.59AVDD345 pKa = 4.16EE346 pKa = 4.51LFLDD350 pKa = 4.56LEE352 pKa = 4.57CMLAFPATVQARR364 pKa = 11.84KK365 pKa = 9.47PEE367 pKa = 4.1CCIMYY372 pKa = 10.3GLEE375 pKa = 4.27TIRR378 pKa = 11.84GSNIKK383 pKa = 10.13LDD385 pKa = 3.71SLAQGFISFATIDD398 pKa = 3.19WSGYY402 pKa = 6.98DD403 pKa = 3.9QRR405 pKa = 11.84LPWFIVRR412 pKa = 11.84AFFFIYY418 pKa = 10.26LPSLLIISHH427 pKa = 6.6GYY429 pKa = 8.34MPTSEE434 pKa = 4.69YY435 pKa = 10.64EE436 pKa = 4.06DD437 pKa = 3.28TSMNISDD444 pKa = 3.51MFTRR448 pKa = 11.84FFNLINFTATWYY460 pKa = 10.72FNMVFLSADD469 pKa = 3.08GFAFRR474 pKa = 11.84RR475 pKa = 11.84QFAGVPSGMLLTQFLDD491 pKa = 3.73SFGNLYY497 pKa = 10.67LIIDD501 pKa = 3.87SLLEE505 pKa = 3.99FGCTYY510 pKa = 11.56DD511 pKa = 5.14DD512 pKa = 4.17IKK514 pKa = 11.59SLMLFIMGDD523 pKa = 3.62DD524 pKa = 3.55NSIFTNWTIDD534 pKa = 3.41KK535 pKa = 9.92LHH537 pKa = 6.93DD538 pKa = 4.49FISFMEE544 pKa = 4.63RR545 pKa = 11.84YY546 pKa = 9.0CLKK549 pKa = 9.56RR550 pKa = 11.84WNMHH554 pKa = 5.18LSKK557 pKa = 9.17TKK559 pKa = 10.71SVITTLRR566 pKa = 11.84SKK568 pKa = 10.66IEE570 pKa = 4.02TLSYY574 pKa = 10.17RR575 pKa = 11.84CNFGKK580 pKa = 9.73PRR582 pKa = 11.84RR583 pKa = 11.84DD584 pKa = 3.36VEE586 pKa = 4.11KK587 pKa = 10.91LIAQLVYY594 pKa = 10.03PEE596 pKa = 5.38HH597 pKa = 6.25GLKK600 pKa = 9.66PQFMSSRR607 pKa = 11.84AIGLAYY613 pKa = 10.1ASCAQDD619 pKa = 3.26STFHH623 pKa = 6.11EE624 pKa = 5.44FCHH627 pKa = 6.49DD628 pKa = 3.66VYY630 pKa = 11.2RR631 pKa = 11.84LYY633 pKa = 11.25LPVADD638 pKa = 5.5LSPAAIRR645 pKa = 11.84NTRR648 pKa = 11.84VWILKK653 pKa = 9.15LLEE656 pKa = 4.17MEE658 pKa = 4.2EE659 pKa = 4.36TEE661 pKa = 4.83ALIPLDD667 pKa = 4.11HH668 pKa = 7.15FPTMSEE674 pKa = 3.84IQHH677 pKa = 6.31LLSYY681 pKa = 9.88YY682 pKa = 10.19HH683 pKa = 6.91GPLRR687 pKa = 11.84PEE689 pKa = 4.17PKK691 pKa = 8.81WNYY694 pKa = 10.04AHH696 pKa = 7.2FPQDD700 pKa = 3.06PDD702 pKa = 3.88FRR704 pKa = 11.84PKK706 pKa = 10.84DD707 pKa = 3.77YY708 pKa = 10.59VTLLDD713 pKa = 3.52YY714 pKa = 10.49MEE716 pKa = 5.31RR717 pKa = 11.84NNISFPEE724 pKa = 4.7LINFTVV730 pKa = 3.1
Molecular weight: 85.78 kDa
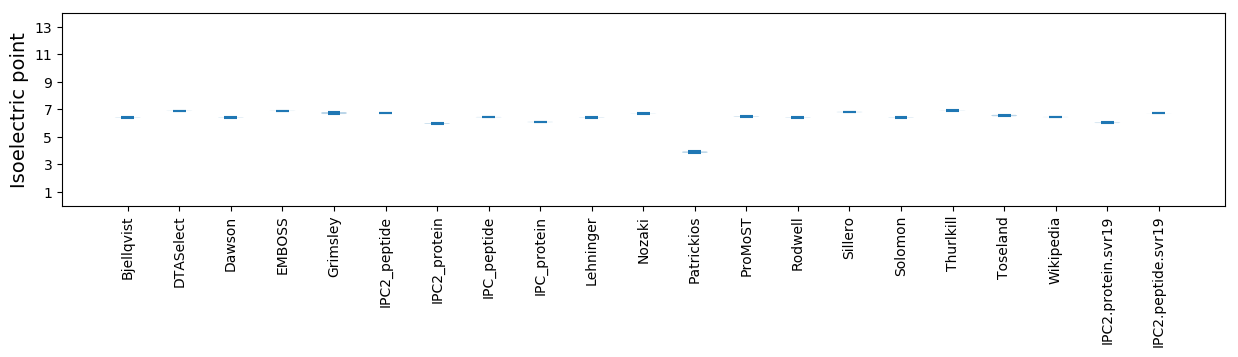
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9Q9X2|Q9Q9X2_9VIRU Putative RNA-dependent RNA polymerase OS=Rhizoctonia solani virus 717 OX=46618 PE=4 SV=1
MM1 pKa = 7.95PSPKK5 pKa = 9.44ISLQDD10 pKa = 3.4RR11 pKa = 11.84LSKK14 pKa = 9.4VTNLNPEE21 pKa = 4.12VKK23 pKa = 9.11FTQGDD28 pKa = 4.23KK29 pKa = 10.96IPDD32 pKa = 3.46RR33 pKa = 11.84AIDD36 pKa = 3.43FHH38 pKa = 7.0EE39 pKa = 4.58VYY41 pKa = 10.31KK42 pKa = 11.13ANSSSKK48 pKa = 9.83IDD50 pKa = 3.49SANVFTIDD58 pKa = 3.43VLPNFVPILCFVIYY72 pKa = 9.8HH73 pKa = 6.38ANVYY77 pKa = 8.89STDD80 pKa = 3.54INKK83 pKa = 9.89HH84 pKa = 4.21GHH86 pKa = 5.99SKK88 pKa = 9.68TSVPTFVSYY97 pKa = 10.97CMSIIYY103 pKa = 9.77GYY105 pKa = 10.83FLLSDD110 pKa = 3.85MYY112 pKa = 11.06VRR114 pKa = 11.84PSPSVFASSWKK125 pKa = 10.53NSTFRR130 pKa = 11.84DD131 pKa = 3.4NFATFLASLPVPEE144 pKa = 4.98FLLPILTSFHH154 pKa = 6.93PFADD158 pKa = 3.88DD159 pKa = 3.11TRR161 pKa = 11.84HH162 pKa = 5.18NVIFVASAAGYY173 pKa = 9.88RR174 pKa = 11.84YY175 pKa = 7.9EE176 pKa = 3.85QHH178 pKa = 6.47FGKK181 pKa = 10.37IIPTLAFYY189 pKa = 10.5NIHH192 pKa = 7.57DD193 pKa = 4.31IVCEE197 pKa = 4.06QPSNAQKK204 pKa = 10.77FDD206 pKa = 3.5ILNALYY212 pKa = 10.67SKK214 pKa = 10.73AIYY217 pKa = 9.97HH218 pKa = 5.7ITATNFNASLANYY231 pKa = 9.28IGYY234 pKa = 10.03GYY236 pKa = 10.63NNAAARR242 pKa = 11.84TNDD245 pKa = 3.64RR246 pKa = 11.84GHH248 pKa = 5.75STGKK252 pKa = 9.99LFQVFEE258 pKa = 4.68SLFSPVLFRR267 pKa = 11.84DD268 pKa = 3.26YY269 pKa = 11.04HH270 pKa = 6.18KK271 pKa = 10.53RR272 pKa = 11.84HH273 pKa = 5.83SLASISLEE281 pKa = 3.84PPTFATSAFNVYY293 pKa = 10.22DD294 pKa = 3.43WLFAATRR301 pKa = 11.84DD302 pKa = 3.6NLAEE306 pKa = 3.98QKK308 pKa = 10.26IVLSSVKK315 pKa = 10.59SFVLDD320 pKa = 3.59SVKK323 pKa = 10.62CSGTLADD330 pKa = 5.28AFTASTGAQITRR342 pKa = 11.84HH343 pKa = 5.93GYY345 pKa = 10.64SLMSLPTAHH354 pKa = 6.91GKK356 pKa = 9.22NLIDD360 pKa = 4.69ANGDD364 pKa = 4.01LISSAKK370 pKa = 10.42LSATDD375 pKa = 3.66RR376 pKa = 11.84QISRR380 pKa = 11.84SARR383 pKa = 11.84DD384 pKa = 3.16IAIEE388 pKa = 4.01IGFLPPIDD396 pKa = 3.91ATPSHH401 pKa = 6.57KK402 pKa = 10.3QPQPQALDD410 pKa = 3.98NKK412 pKa = 9.68DD413 pKa = 3.44QPIPDD418 pKa = 3.65SHH420 pKa = 8.22VSNASWPWNLLHH432 pKa = 6.52STLNPPTAPKK442 pKa = 10.42LSDD445 pKa = 3.74VIQVPSPEE453 pKa = 4.17TEE455 pKa = 3.9FEE457 pKa = 4.02IFDD460 pKa = 4.21SDD462 pKa = 3.31RR463 pKa = 11.84HH464 pKa = 5.95HH465 pKa = 6.45YY466 pKa = 10.18PPVRR470 pKa = 11.84VLNFPDD476 pKa = 4.85DD477 pKa = 4.11STVDD481 pKa = 3.21AHH483 pKa = 7.01LATLTGMVIEE493 pKa = 4.61SFEE496 pKa = 4.1IDD498 pKa = 3.7GSVVAHH504 pKa = 6.95PDD506 pKa = 2.93AHH508 pKa = 6.91LSNGVDD514 pKa = 3.48NTWFAVSAISLRR526 pKa = 11.84YY527 pKa = 8.35VYY529 pKa = 10.32QYY531 pKa = 9.42NTLQNGNASPIRR543 pKa = 11.84ARR545 pKa = 11.84SRR547 pKa = 11.84VFRR550 pKa = 11.84PTQGAHH556 pKa = 7.14PIATLFVDD564 pKa = 4.24RR565 pKa = 11.84TQINLPRR572 pKa = 11.84PAPYY576 pKa = 10.35LYY578 pKa = 10.62DD579 pKa = 3.3QLVNNTLPGLTIVQNATWIAKK600 pKa = 8.82VKK602 pKa = 10.66SFLGSNVSHH611 pKa = 7.28RR612 pKa = 11.84LRR614 pKa = 11.84RR615 pKa = 11.84TEE617 pKa = 3.49NTYY620 pKa = 10.73PGFDD624 pKa = 4.06DD625 pKa = 4.25RR626 pKa = 11.84DD627 pKa = 3.55IYY629 pKa = 10.95VWSPYY634 pKa = 10.46SYY636 pKa = 10.51TPVSPRR642 pKa = 11.84DD643 pKa = 3.68DD644 pKa = 4.38ADD646 pKa = 3.02IDD648 pKa = 3.74YY649 pKa = 10.62SEE651 pKa = 4.14QVTYY655 pKa = 10.82FLTNLRR661 pKa = 11.84TIFGTRR667 pKa = 11.84NRR669 pKa = 11.84MIEE672 pKa = 4.04VQNSLEE678 pKa = 4.04VMPVMM683 pKa = 5.69
MM1 pKa = 7.95PSPKK5 pKa = 9.44ISLQDD10 pKa = 3.4RR11 pKa = 11.84LSKK14 pKa = 9.4VTNLNPEE21 pKa = 4.12VKK23 pKa = 9.11FTQGDD28 pKa = 4.23KK29 pKa = 10.96IPDD32 pKa = 3.46RR33 pKa = 11.84AIDD36 pKa = 3.43FHH38 pKa = 7.0EE39 pKa = 4.58VYY41 pKa = 10.31KK42 pKa = 11.13ANSSSKK48 pKa = 9.83IDD50 pKa = 3.49SANVFTIDD58 pKa = 3.43VLPNFVPILCFVIYY72 pKa = 9.8HH73 pKa = 6.38ANVYY77 pKa = 8.89STDD80 pKa = 3.54INKK83 pKa = 9.89HH84 pKa = 4.21GHH86 pKa = 5.99SKK88 pKa = 9.68TSVPTFVSYY97 pKa = 10.97CMSIIYY103 pKa = 9.77GYY105 pKa = 10.83FLLSDD110 pKa = 3.85MYY112 pKa = 11.06VRR114 pKa = 11.84PSPSVFASSWKK125 pKa = 10.53NSTFRR130 pKa = 11.84DD131 pKa = 3.4NFATFLASLPVPEE144 pKa = 4.98FLLPILTSFHH154 pKa = 6.93PFADD158 pKa = 3.88DD159 pKa = 3.11TRR161 pKa = 11.84HH162 pKa = 5.18NVIFVASAAGYY173 pKa = 9.88RR174 pKa = 11.84YY175 pKa = 7.9EE176 pKa = 3.85QHH178 pKa = 6.47FGKK181 pKa = 10.37IIPTLAFYY189 pKa = 10.5NIHH192 pKa = 7.57DD193 pKa = 4.31IVCEE197 pKa = 4.06QPSNAQKK204 pKa = 10.77FDD206 pKa = 3.5ILNALYY212 pKa = 10.67SKK214 pKa = 10.73AIYY217 pKa = 9.97HH218 pKa = 5.7ITATNFNASLANYY231 pKa = 9.28IGYY234 pKa = 10.03GYY236 pKa = 10.63NNAAARR242 pKa = 11.84TNDD245 pKa = 3.64RR246 pKa = 11.84GHH248 pKa = 5.75STGKK252 pKa = 9.99LFQVFEE258 pKa = 4.68SLFSPVLFRR267 pKa = 11.84DD268 pKa = 3.26YY269 pKa = 11.04HH270 pKa = 6.18KK271 pKa = 10.53RR272 pKa = 11.84HH273 pKa = 5.83SLASISLEE281 pKa = 3.84PPTFATSAFNVYY293 pKa = 10.22DD294 pKa = 3.43WLFAATRR301 pKa = 11.84DD302 pKa = 3.6NLAEE306 pKa = 3.98QKK308 pKa = 10.26IVLSSVKK315 pKa = 10.59SFVLDD320 pKa = 3.59SVKK323 pKa = 10.62CSGTLADD330 pKa = 5.28AFTASTGAQITRR342 pKa = 11.84HH343 pKa = 5.93GYY345 pKa = 10.64SLMSLPTAHH354 pKa = 6.91GKK356 pKa = 9.22NLIDD360 pKa = 4.69ANGDD364 pKa = 4.01LISSAKK370 pKa = 10.42LSATDD375 pKa = 3.66RR376 pKa = 11.84QISRR380 pKa = 11.84SARR383 pKa = 11.84DD384 pKa = 3.16IAIEE388 pKa = 4.01IGFLPPIDD396 pKa = 3.91ATPSHH401 pKa = 6.57KK402 pKa = 10.3QPQPQALDD410 pKa = 3.98NKK412 pKa = 9.68DD413 pKa = 3.44QPIPDD418 pKa = 3.65SHH420 pKa = 8.22VSNASWPWNLLHH432 pKa = 6.52STLNPPTAPKK442 pKa = 10.42LSDD445 pKa = 3.74VIQVPSPEE453 pKa = 4.17TEE455 pKa = 3.9FEE457 pKa = 4.02IFDD460 pKa = 4.21SDD462 pKa = 3.31RR463 pKa = 11.84HH464 pKa = 5.95HH465 pKa = 6.45YY466 pKa = 10.18PPVRR470 pKa = 11.84VLNFPDD476 pKa = 4.85DD477 pKa = 4.11STVDD481 pKa = 3.21AHH483 pKa = 7.01LATLTGMVIEE493 pKa = 4.61SFEE496 pKa = 4.1IDD498 pKa = 3.7GSVVAHH504 pKa = 6.95PDD506 pKa = 2.93AHH508 pKa = 6.91LSNGVDD514 pKa = 3.48NTWFAVSAISLRR526 pKa = 11.84YY527 pKa = 8.35VYY529 pKa = 10.32QYY531 pKa = 9.42NTLQNGNASPIRR543 pKa = 11.84ARR545 pKa = 11.84SRR547 pKa = 11.84VFRR550 pKa = 11.84PTQGAHH556 pKa = 7.14PIATLFVDD564 pKa = 4.24RR565 pKa = 11.84TQINLPRR572 pKa = 11.84PAPYY576 pKa = 10.35LYY578 pKa = 10.62DD579 pKa = 3.3QLVNNTLPGLTIVQNATWIAKK600 pKa = 8.82VKK602 pKa = 10.66SFLGSNVSHH611 pKa = 7.28RR612 pKa = 11.84LRR614 pKa = 11.84RR615 pKa = 11.84TEE617 pKa = 3.49NTYY620 pKa = 10.73PGFDD624 pKa = 4.06DD625 pKa = 4.25RR626 pKa = 11.84DD627 pKa = 3.55IYY629 pKa = 10.95VWSPYY634 pKa = 10.46SYY636 pKa = 10.51TPVSPRR642 pKa = 11.84DD643 pKa = 3.68DD644 pKa = 4.38ADD646 pKa = 3.02IDD648 pKa = 3.74YY649 pKa = 10.62SEE651 pKa = 4.14QVTYY655 pKa = 10.82FLTNLRR661 pKa = 11.84TIFGTRR667 pKa = 11.84NRR669 pKa = 11.84MIEE672 pKa = 4.04VQNSLEE678 pKa = 4.04VMPVMM683 pKa = 5.69
Molecular weight: 76.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1413 |
683 |
730 |
706.5 |
81.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.369 ± 1.205 | 1.062 ± 0.341 |
6.723 ± 0.096 | 3.751 ± 0.799 |
7.077 ± 0.874 | 3.539 ± 0.018 |
3.468 ± 0.033 | 6.653 ± 0.046 |
4.176 ± 0.474 | 8.846 ± 0.673 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.265 ± 0.783 | 5.449 ± 0.501 |
6.723 ± 0.218 | 2.76 ± 0.33 |
5.308 ± 0.446 | 8.209 ± 1.251 |
6.582 ± 0.11 | 4.883 ± 1.431 |
1.062 ± 0.026 | 5.096 ± 0.504 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
