
Streptococcus satellite phage Javan86
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
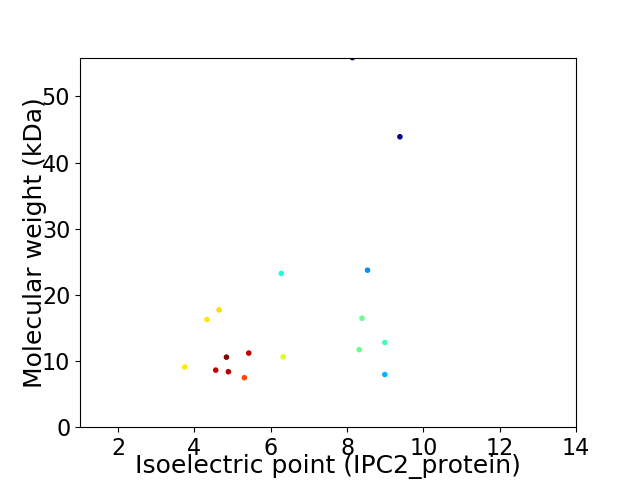
Virtual 2D-PAGE plot for 17 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZXQ5|A0A4D5ZXQ5_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan86 OX=2558865 GN=JavanS86_0017 PE=4 SV=1
MM1 pKa = 7.63TDD3 pKa = 3.77KK4 pKa = 10.59KK5 pKa = 9.49TIDD8 pKa = 3.59LNNYY12 pKa = 9.36GITDD16 pKa = 3.58KK17 pKa = 11.55DD18 pKa = 3.64GDD20 pKa = 4.29DD21 pKa = 3.33VTFYY25 pKa = 11.09QIANFYY31 pKa = 11.25NSFLEE36 pKa = 5.17VADD39 pKa = 3.9HH40 pKa = 6.74WLDD43 pKa = 4.03AEE45 pKa = 4.43LTDD48 pKa = 4.17GQAWRR53 pKa = 11.84YY54 pKa = 7.52MMQIYY59 pKa = 10.59AEE61 pKa = 4.19LFLYY65 pKa = 10.29PEE67 pKa = 5.16DD68 pKa = 4.37EE69 pKa = 4.37PASTEE74 pKa = 3.87EE75 pKa = 4.09TTAA78 pKa = 3.47
MM1 pKa = 7.63TDD3 pKa = 3.77KK4 pKa = 10.59KK5 pKa = 9.49TIDD8 pKa = 3.59LNNYY12 pKa = 9.36GITDD16 pKa = 3.58KK17 pKa = 11.55DD18 pKa = 3.64GDD20 pKa = 4.29DD21 pKa = 3.33VTFYY25 pKa = 11.09QIANFYY31 pKa = 11.25NSFLEE36 pKa = 5.17VADD39 pKa = 3.9HH40 pKa = 6.74WLDD43 pKa = 4.03AEE45 pKa = 4.43LTDD48 pKa = 4.17GQAWRR53 pKa = 11.84YY54 pKa = 7.52MMQIYY59 pKa = 10.59AEE61 pKa = 4.19LFLYY65 pKa = 10.29PEE67 pKa = 5.16DD68 pKa = 4.37EE69 pKa = 4.37PASTEE74 pKa = 3.87EE75 pKa = 4.09TTAA78 pKa = 3.47
Molecular weight: 9.1 kDa
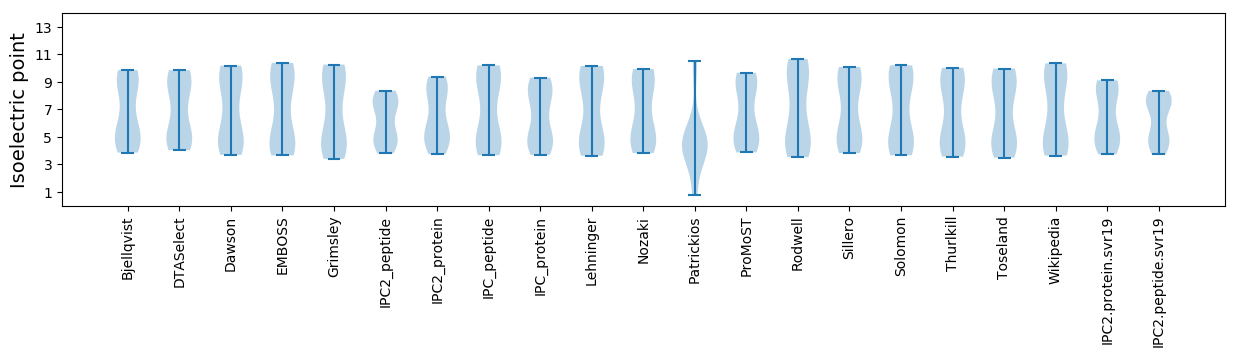
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZWR1|A0A4D5ZWR1_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan86 OX=2558865 GN=JavanS86_0009 PE=4 SV=1
MM1 pKa = 7.65TIKK4 pKa = 10.49QFTKK8 pKa = 10.54KK9 pKa = 10.34NGEE12 pKa = 3.88TVYY15 pKa = 10.45RR16 pKa = 11.84QRR18 pKa = 11.84VYY20 pKa = 11.13LGTDD24 pKa = 2.68SMTGKK29 pKa = 9.73QVQVTITGRR38 pKa = 11.84TKK40 pKa = 10.87SEE42 pKa = 3.92VSQKK46 pKa = 10.64AIKK49 pKa = 10.18KK50 pKa = 9.95LADD53 pKa = 3.75FQANGSTVLKK63 pKa = 8.65MQQVNNYY70 pKa = 9.31SDD72 pKa = 5.97LVTLWLSSYY81 pKa = 10.83KK82 pKa = 10.06LTVKK86 pKa = 9.91PQTYY90 pKa = 8.71RR91 pKa = 11.84HH92 pKa = 5.32TEE94 pKa = 3.64ITIHH98 pKa = 5.86KK99 pKa = 9.4HH100 pKa = 5.69LIPFFGTMKK109 pKa = 10.39LEE111 pKa = 4.65KK112 pKa = 9.79ISMPFVQYY120 pKa = 9.53FANSISSSGLKK131 pKa = 9.53EE132 pKa = 3.99YY133 pKa = 10.95KK134 pKa = 10.24PIIALNKK141 pKa = 10.1RR142 pKa = 11.84ILQYY146 pKa = 10.9AVHH149 pKa = 6.14LQIITRR155 pKa = 11.84NPADD159 pKa = 4.25NIIVPCMRR167 pKa = 11.84QEE169 pKa = 4.16EE170 pKa = 4.64KK171 pKa = 10.57NLKK174 pKa = 10.08YY175 pKa = 10.74LDD177 pKa = 4.47DD178 pKa = 4.21SQLKK182 pKa = 9.46QFIGYY187 pKa = 9.78LDD189 pKa = 4.16SLPDD193 pKa = 3.58NYY195 pKa = 10.77KK196 pKa = 10.4NNYY199 pKa = 8.48NRR201 pKa = 11.84VLYY204 pKa = 10.36QFLLATGLRR213 pKa = 11.84IGEE216 pKa = 4.54AIALEE221 pKa = 4.14WSDD224 pKa = 3.66IDD226 pKa = 5.51LEE228 pKa = 4.47NGTINITKK236 pKa = 8.95TFNPVINKK244 pKa = 9.87LSTPKK249 pKa = 9.78TKK251 pKa = 10.37AGKK254 pKa = 9.83RR255 pKa = 11.84VISIDD260 pKa = 3.58RR261 pKa = 11.84KK262 pKa = 9.09SLLMLRR268 pKa = 11.84LYY270 pKa = 10.39KK271 pKa = 10.22VRR273 pKa = 11.84QEE275 pKa = 4.08QKK277 pKa = 8.68FKK279 pKa = 11.4EE280 pKa = 4.24MISPYY285 pKa = 10.42GNYY288 pKa = 9.53VFSNGLAPYY297 pKa = 9.29PFRR300 pKa = 11.84NNLQRR305 pKa = 11.84MLDD308 pKa = 3.56NHH310 pKa = 6.67LEE312 pKa = 4.18RR313 pKa = 11.84AGLPRR318 pKa = 11.84FTFHH322 pKa = 7.9AFRR325 pKa = 11.84HH326 pKa = 4.69THH328 pKa = 6.92ASLLLNAGISYY339 pKa = 10.65KK340 pKa = 10.3EE341 pKa = 3.81LQHH344 pKa = 7.01RR345 pKa = 11.84LGHH348 pKa = 5.9SKK350 pKa = 10.3ISMTLDD356 pKa = 3.44VYY358 pKa = 11.48SHH360 pKa = 6.96LSKK363 pKa = 10.93SKK365 pKa = 9.49EE366 pKa = 4.09KK367 pKa = 10.76EE368 pKa = 3.62AVTYY372 pKa = 9.03YY373 pKa = 10.81EE374 pKa = 4.2KK375 pKa = 11.2ALGKK379 pKa = 10.25LL380 pKa = 3.53
MM1 pKa = 7.65TIKK4 pKa = 10.49QFTKK8 pKa = 10.54KK9 pKa = 10.34NGEE12 pKa = 3.88TVYY15 pKa = 10.45RR16 pKa = 11.84QRR18 pKa = 11.84VYY20 pKa = 11.13LGTDD24 pKa = 2.68SMTGKK29 pKa = 9.73QVQVTITGRR38 pKa = 11.84TKK40 pKa = 10.87SEE42 pKa = 3.92VSQKK46 pKa = 10.64AIKK49 pKa = 10.18KK50 pKa = 9.95LADD53 pKa = 3.75FQANGSTVLKK63 pKa = 8.65MQQVNNYY70 pKa = 9.31SDD72 pKa = 5.97LVTLWLSSYY81 pKa = 10.83KK82 pKa = 10.06LTVKK86 pKa = 9.91PQTYY90 pKa = 8.71RR91 pKa = 11.84HH92 pKa = 5.32TEE94 pKa = 3.64ITIHH98 pKa = 5.86KK99 pKa = 9.4HH100 pKa = 5.69LIPFFGTMKK109 pKa = 10.39LEE111 pKa = 4.65KK112 pKa = 9.79ISMPFVQYY120 pKa = 9.53FANSISSSGLKK131 pKa = 9.53EE132 pKa = 3.99YY133 pKa = 10.95KK134 pKa = 10.24PIIALNKK141 pKa = 10.1RR142 pKa = 11.84ILQYY146 pKa = 10.9AVHH149 pKa = 6.14LQIITRR155 pKa = 11.84NPADD159 pKa = 4.25NIIVPCMRR167 pKa = 11.84QEE169 pKa = 4.16EE170 pKa = 4.64KK171 pKa = 10.57NLKK174 pKa = 10.08YY175 pKa = 10.74LDD177 pKa = 4.47DD178 pKa = 4.21SQLKK182 pKa = 9.46QFIGYY187 pKa = 9.78LDD189 pKa = 4.16SLPDD193 pKa = 3.58NYY195 pKa = 10.77KK196 pKa = 10.4NNYY199 pKa = 8.48NRR201 pKa = 11.84VLYY204 pKa = 10.36QFLLATGLRR213 pKa = 11.84IGEE216 pKa = 4.54AIALEE221 pKa = 4.14WSDD224 pKa = 3.66IDD226 pKa = 5.51LEE228 pKa = 4.47NGTINITKK236 pKa = 8.95TFNPVINKK244 pKa = 9.87LSTPKK249 pKa = 9.78TKK251 pKa = 10.37AGKK254 pKa = 9.83RR255 pKa = 11.84VISIDD260 pKa = 3.58RR261 pKa = 11.84KK262 pKa = 9.09SLLMLRR268 pKa = 11.84LYY270 pKa = 10.39KK271 pKa = 10.22VRR273 pKa = 11.84QEE275 pKa = 4.08QKK277 pKa = 8.68FKK279 pKa = 11.4EE280 pKa = 4.24MISPYY285 pKa = 10.42GNYY288 pKa = 9.53VFSNGLAPYY297 pKa = 9.29PFRR300 pKa = 11.84NNLQRR305 pKa = 11.84MLDD308 pKa = 3.56NHH310 pKa = 6.67LEE312 pKa = 4.18RR313 pKa = 11.84AGLPRR318 pKa = 11.84FTFHH322 pKa = 7.9AFRR325 pKa = 11.84HH326 pKa = 4.69THH328 pKa = 6.92ASLLLNAGISYY339 pKa = 10.65KK340 pKa = 10.3EE341 pKa = 3.81LQHH344 pKa = 7.01RR345 pKa = 11.84LGHH348 pKa = 5.9SKK350 pKa = 10.3ISMTLDD356 pKa = 3.44VYY358 pKa = 11.48SHH360 pKa = 6.96LSKK363 pKa = 10.93SKK365 pKa = 9.49EE366 pKa = 4.09KK367 pKa = 10.76EE368 pKa = 3.62AVTYY372 pKa = 9.03YY373 pKa = 10.81EE374 pKa = 4.2KK375 pKa = 11.2ALGKK379 pKa = 10.25LL380 pKa = 3.53
Molecular weight: 43.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2547 |
65 |
475 |
149.8 |
17.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.575 ± 0.527 | 0.51 ± 0.153 |
5.693 ± 0.578 | 8.638 ± 0.714 |
3.691 ± 0.305 | 4.947 ± 0.422 |
1.413 ± 0.316 | 6.243 ± 0.396 |
8.716 ± 0.5 | 10.561 ± 0.583 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.866 ± 0.415 | 5.575 ± 0.542 |
2.67 ± 0.325 | 4.476 ± 0.491 |
5.457 ± 0.62 | 5.732 ± 0.29 |
6.243 ± 0.373 | 4.751 ± 0.353 |
0.942 ± 0.216 | 5.3 ± 0.301 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
