
Suhomyces tanzawaensis NRRL Y-17324
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Debaryomycetaceae; Suhomyces; Suhomyces tanzawaensis
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
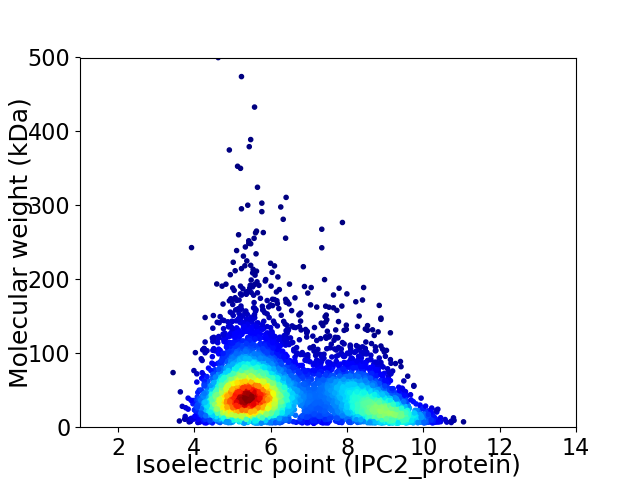
Virtual 2D-PAGE plot for 5882 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E4SR02|A0A1E4SR02_9ASCO Coatomer subunit beta' OS=Suhomyces tanzawaensis NRRL Y-17324 OX=984487 GN=CANTADRAFT_46178 PE=3 SV=1
GG1 pKa = 7.44SYY3 pKa = 10.25SANLTNEE10 pKa = 3.87NVFYY14 pKa = 7.85TTNITVGSEE23 pKa = 3.75KK24 pKa = 10.65KK25 pKa = 10.41QILVDD30 pKa = 4.57LDD32 pKa = 3.9TGSSDD37 pKa = 4.18LWVLSANGTDD47 pKa = 4.24GFASDD52 pKa = 3.61TAEE55 pKa = 4.12LQSNGYY61 pKa = 10.07LDD63 pKa = 4.46AEE65 pKa = 4.66KK66 pKa = 9.99STSLKK71 pKa = 9.43TDD73 pKa = 3.03NNEE76 pKa = 3.7FFITYY81 pKa = 10.29GDD83 pKa = 3.3NTFAYY88 pKa = 9.69GEE90 pKa = 4.38YY91 pKa = 9.07VTDD94 pKa = 5.64SIAIPNGPSIDD105 pKa = 3.77VEE107 pKa = 4.29FGLAFTTVNPFGIFGIGFDD126 pKa = 4.03LNEE129 pKa = 4.57AGFSSGAAPIYY140 pKa = 10.49NNYY143 pKa = 8.91PATLKK148 pKa = 11.03KK149 pKa = 10.58NGLIGKK155 pKa = 8.33NAYY158 pKa = 9.79SLYY161 pKa = 10.54LNSPNATTGSVIFGGYY177 pKa = 7.35DD178 pKa = 3.19TKK180 pKa = 11.22KK181 pKa = 10.29VDD183 pKa = 3.53GSFATLQTVNDD194 pKa = 3.56QVLAINVDD202 pKa = 3.78SVTVGDD208 pKa = 4.63NEE210 pKa = 4.06IQYY213 pKa = 10.51GSPALLDD220 pKa = 3.59SGTTITYY227 pKa = 10.05LPRR230 pKa = 11.84NVYY233 pKa = 10.1QGIIQNLPGSNTPYY247 pKa = 10.31GWRR250 pKa = 11.84ITTDD254 pKa = 2.88EE255 pKa = 4.37AQGHH259 pKa = 7.01DD260 pKa = 3.08ITYY263 pKa = 10.16KK264 pKa = 10.77FNDD267 pKa = 3.46VTVKK271 pKa = 10.59VPVSSLVGPTYY282 pKa = 10.84DD283 pKa = 4.7DD284 pKa = 3.1QGKK287 pKa = 9.25LLDD290 pKa = 3.93YY291 pKa = 10.7TSLQVSIGGSILGDD305 pKa = 3.16NFLRR309 pKa = 11.84YY310 pKa = 9.86AYY312 pKa = 10.56VIFNLDD318 pKa = 3.39DD319 pKa = 3.57QKK321 pKa = 11.53ISVAQAKK328 pKa = 7.99FTDD331 pKa = 4.31DD332 pKa = 4.4SDD334 pKa = 3.6IQAII338 pKa = 3.83
GG1 pKa = 7.44SYY3 pKa = 10.25SANLTNEE10 pKa = 3.87NVFYY14 pKa = 7.85TTNITVGSEE23 pKa = 3.75KK24 pKa = 10.65KK25 pKa = 10.41QILVDD30 pKa = 4.57LDD32 pKa = 3.9TGSSDD37 pKa = 4.18LWVLSANGTDD47 pKa = 4.24GFASDD52 pKa = 3.61TAEE55 pKa = 4.12LQSNGYY61 pKa = 10.07LDD63 pKa = 4.46AEE65 pKa = 4.66KK66 pKa = 9.99STSLKK71 pKa = 9.43TDD73 pKa = 3.03NNEE76 pKa = 3.7FFITYY81 pKa = 10.29GDD83 pKa = 3.3NTFAYY88 pKa = 9.69GEE90 pKa = 4.38YY91 pKa = 9.07VTDD94 pKa = 5.64SIAIPNGPSIDD105 pKa = 3.77VEE107 pKa = 4.29FGLAFTTVNPFGIFGIGFDD126 pKa = 4.03LNEE129 pKa = 4.57AGFSSGAAPIYY140 pKa = 10.49NNYY143 pKa = 8.91PATLKK148 pKa = 11.03KK149 pKa = 10.58NGLIGKK155 pKa = 8.33NAYY158 pKa = 9.79SLYY161 pKa = 10.54LNSPNATTGSVIFGGYY177 pKa = 7.35DD178 pKa = 3.19TKK180 pKa = 11.22KK181 pKa = 10.29VDD183 pKa = 3.53GSFATLQTVNDD194 pKa = 3.56QVLAINVDD202 pKa = 3.78SVTVGDD208 pKa = 4.63NEE210 pKa = 4.06IQYY213 pKa = 10.51GSPALLDD220 pKa = 3.59SGTTITYY227 pKa = 10.05LPRR230 pKa = 11.84NVYY233 pKa = 10.1QGIIQNLPGSNTPYY247 pKa = 10.31GWRR250 pKa = 11.84ITTDD254 pKa = 2.88EE255 pKa = 4.37AQGHH259 pKa = 7.01DD260 pKa = 3.08ITYY263 pKa = 10.16KK264 pKa = 10.77FNDD267 pKa = 3.46VTVKK271 pKa = 10.59VPVSSLVGPTYY282 pKa = 10.84DD283 pKa = 4.7DD284 pKa = 3.1QGKK287 pKa = 9.25LLDD290 pKa = 3.93YY291 pKa = 10.7TSLQVSIGGSILGDD305 pKa = 3.16NFLRR309 pKa = 11.84YY310 pKa = 9.86AYY312 pKa = 10.56VIFNLDD318 pKa = 3.39DD319 pKa = 3.57QKK321 pKa = 11.53ISVAQAKK328 pKa = 7.99FTDD331 pKa = 4.31DD332 pKa = 4.4SDD334 pKa = 3.6IQAII338 pKa = 3.83
Molecular weight: 36.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E4SH60|A0A1E4SH60_9ASCO DUF676 domain-containing protein OS=Suhomyces tanzawaensis NRRL Y-17324 OX=984487 GN=CANTADRAFT_22712 PE=3 SV=1
MM1 pKa = 7.4TPATQQWWGSPSPSPRR17 pKa = 11.84PNRR20 pKa = 11.84RR21 pKa = 11.84GARR24 pKa = 11.84VRR26 pKa = 11.84WGRR29 pKa = 11.84TNGSPPAAIPLCSPPPGSKK48 pKa = 10.58GPPTFRR54 pKa = 11.84KK55 pKa = 9.56RR56 pKa = 11.84QVPLSRR62 pKa = 11.84GGAKK66 pKa = 9.76SEE68 pKa = 4.11PLLSRR73 pKa = 11.84GGMLRR78 pKa = 11.84ASPRR82 pKa = 11.84RR83 pKa = 11.84LRR85 pKa = 3.78
MM1 pKa = 7.4TPATQQWWGSPSPSPRR17 pKa = 11.84PNRR20 pKa = 11.84RR21 pKa = 11.84GARR24 pKa = 11.84VRR26 pKa = 11.84WGRR29 pKa = 11.84TNGSPPAAIPLCSPPPGSKK48 pKa = 10.58GPPTFRR54 pKa = 11.84KK55 pKa = 9.56RR56 pKa = 11.84QVPLSRR62 pKa = 11.84GGAKK66 pKa = 9.76SEE68 pKa = 4.11PLLSRR73 pKa = 11.84GGMLRR78 pKa = 11.84ASPRR82 pKa = 11.84RR83 pKa = 11.84LRR85 pKa = 3.78
Molecular weight: 9.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2688497 |
50 |
4399 |
457.1 |
51.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.097 ± 0.034 | 1.048 ± 0.011 |
5.844 ± 0.025 | 6.541 ± 0.04 |
4.399 ± 0.02 | 5.425 ± 0.028 |
2.296 ± 0.014 | 6.376 ± 0.027 |
6.925 ± 0.034 | 9.938 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.803 ± 0.011 | 5.494 ± 0.028 |
4.723 ± 0.029 | 4.031 ± 0.025 |
4.26 ± 0.022 | 8.704 ± 0.041 |
5.595 ± 0.018 | 6.0 ± 0.021 |
1.041 ± 0.01 | 3.454 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
