
TM7 phylum sp. oral taxon 349
Taxonomy: cellular organisms; Bacteria; Bacteria incertae sedis; Bacteria candidate phyla; Candidatus Saccharibacteria
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
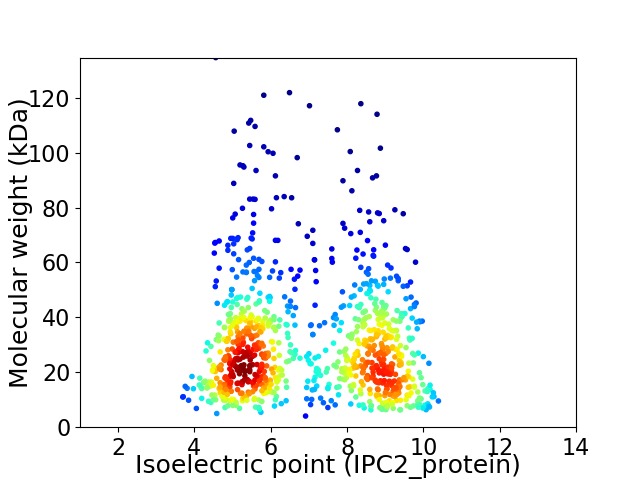
Virtual 2D-PAGE plot for 963 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A563CXA7|A0A563CXA7_9BACT Uncharacterized protein OS=TM7 phylum sp. oral taxon 349 OX=713051 GN=EUA66_02615 PE=4 SV=1
MM1 pKa = 7.58EE2 pKa = 5.9LNPGEE7 pKa = 4.21RR8 pKa = 11.84YY9 pKa = 9.08EE10 pKa = 4.3VAFDD14 pKa = 3.89SYY16 pKa = 11.06TGNLAVGEE24 pKa = 4.25TVEE27 pKa = 5.95FYY29 pKa = 11.36DD30 pKa = 5.13NNDD33 pKa = 4.51LICVEE38 pKa = 5.05LEE40 pKa = 3.97NLDD43 pKa = 4.49DD44 pKa = 4.67SMGNVIRR51 pKa = 11.84AIGGVAGQEE60 pKa = 4.28IIMSSGLFSCDD71 pKa = 3.18HH72 pKa = 6.9GDD74 pKa = 3.18GFAIFKK80 pKa = 10.28ARR82 pKa = 11.84HH83 pKa = 6.55DD84 pKa = 5.14DD85 pKa = 3.64IQDD88 pKa = 3.31VSS90 pKa = 3.48
MM1 pKa = 7.58EE2 pKa = 5.9LNPGEE7 pKa = 4.21RR8 pKa = 11.84YY9 pKa = 9.08EE10 pKa = 4.3VAFDD14 pKa = 3.89SYY16 pKa = 11.06TGNLAVGEE24 pKa = 4.25TVEE27 pKa = 5.95FYY29 pKa = 11.36DD30 pKa = 5.13NNDD33 pKa = 4.51LICVEE38 pKa = 5.05LEE40 pKa = 3.97NLDD43 pKa = 4.49DD44 pKa = 4.67SMGNVIRR51 pKa = 11.84AIGGVAGQEE60 pKa = 4.28IIMSSGLFSCDD71 pKa = 3.18HH72 pKa = 6.9GDD74 pKa = 3.18GFAIFKK80 pKa = 10.28ARR82 pKa = 11.84HH83 pKa = 6.55DD84 pKa = 5.14DD85 pKa = 3.64IQDD88 pKa = 3.31VSS90 pKa = 3.48
Molecular weight: 9.82 kDa
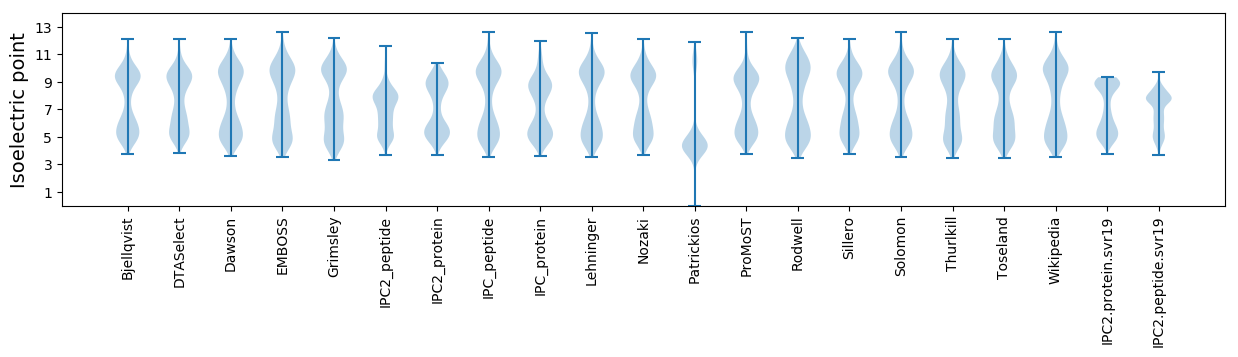
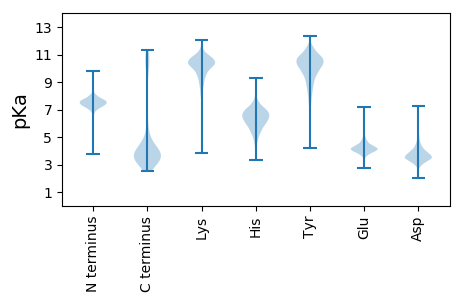
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A563D216|A0A563D216_9BACT Uncharacterized protein OS=TM7 phylum sp. oral taxon 349 OX=713051 GN=EUA66_01090 PE=4 SV=1
MM1 pKa = 7.34NGTWRR6 pKa = 11.84WQEE9 pKa = 3.99LAWDD13 pKa = 4.19SPATPAVGAAALEE26 pKa = 4.17RR27 pKa = 11.84LTTAVEE33 pKa = 3.99VGPVVLEE40 pKa = 3.71LRR42 pKa = 11.84AWAGGAYY49 pKa = 8.59WLAGCQPGRR58 pKa = 11.84AVEE61 pKa = 4.28LKK63 pKa = 10.46QLLGHH68 pKa = 6.71HH69 pKa = 6.6LPARR73 pKa = 11.84LHH75 pKa = 5.93RR76 pKa = 11.84PQRR79 pKa = 11.84QRR81 pKa = 11.84LAVGQAVRR89 pKa = 11.84LQVRR93 pKa = 11.84GNHH96 pKa = 5.39VSTSADD102 pKa = 2.7RR103 pKa = 11.84VVAATRR109 pKa = 11.84ALYY112 pKa = 10.42ACLSGLHH119 pKa = 6.43EE120 pKa = 4.39NQQAVVQLLIGRR132 pKa = 11.84RR133 pKa = 11.84LPSSSFTGPPAPGWRR148 pKa = 11.84EE149 pKa = 3.79LFLGQPAPRR158 pKa = 11.84SPEE161 pKa = 3.72RR162 pKa = 11.84PTRR165 pKa = 11.84TTSEE169 pKa = 3.7QHH171 pKa = 5.69GAAACLRR178 pKa = 11.84LGHH181 pKa = 6.72TGTPAQAHH189 pKa = 4.75QLFSRR194 pKa = 11.84LLGALRR200 pKa = 11.84IVEE203 pKa = 4.26TPRR206 pKa = 11.84ARR208 pKa = 11.84FRR210 pKa = 11.84LSAEE214 pKa = 3.8APDD217 pKa = 3.51RR218 pKa = 11.84LNRR221 pKa = 11.84VSRR224 pKa = 11.84PWAWPLRR231 pKa = 11.84LRR233 pKa = 11.84SGQLTALCGWPIGEE247 pKa = 4.84PPLPLLGDD255 pKa = 3.54LHH257 pKa = 6.83PRR259 pKa = 11.84LLPPPASLVRR269 pKa = 11.84SEE271 pKa = 4.48RR272 pKa = 11.84VIGLASAPGCRR283 pKa = 11.84QPVAIPIRR291 pKa = 11.84DD292 pKa = 3.75AVFHH296 pKa = 5.55THH298 pKa = 7.12LLGPTGTGKK307 pKa = 8.49STVLLSLALADD318 pKa = 4.13VEE320 pKa = 4.12AHH322 pKa = 6.42RR323 pKa = 11.84AVLLLDD329 pKa = 3.97PKK331 pKa = 11.28GDD333 pKa = 3.71LATDD337 pKa = 3.93LLARR341 pKa = 11.84VPEE344 pKa = 4.47GRR346 pKa = 11.84HH347 pKa = 5.18EE348 pKa = 4.35DD349 pKa = 3.68VVVIDD354 pKa = 4.25PTNPAPVGLNPLAGPARR371 pKa = 11.84LAPVTADD378 pKa = 3.31SLLGTLAALFHH389 pKa = 5.8EE390 pKa = 4.71HH391 pKa = 6.02WGVRR395 pKa = 11.84TADD398 pKa = 3.6VLNAALLTLARR409 pKa = 11.84TPGANLLWLPPLLTDD424 pKa = 3.41PAFRR428 pKa = 11.84RR429 pKa = 11.84RR430 pKa = 11.84VLDD433 pKa = 4.38DD434 pKa = 3.31QDD436 pKa = 5.23DD437 pKa = 4.09PLGTGAFWAAYY448 pKa = 6.69EE449 pKa = 4.15AKK451 pKa = 10.14RR452 pKa = 11.84PQAQAVEE459 pKa = 4.14IAPVLNKK466 pKa = 9.82LRR468 pKa = 11.84QLILRR473 pKa = 11.84PHH475 pKa = 6.2LRR477 pKa = 11.84AVLGQSAPRR486 pKa = 11.84FTVEE490 pKa = 3.89EE491 pKa = 4.29LFTRR495 pKa = 11.84KK496 pKa = 9.96RR497 pKa = 11.84IVVVNLNRR505 pKa = 11.84GLLGAEE511 pKa = 4.34AARR514 pKa = 11.84LLGSLLVGQLWSRR527 pKa = 11.84LLARR531 pKa = 11.84QALPPEE537 pKa = 4.28RR538 pKa = 11.84RR539 pKa = 11.84HH540 pKa = 5.23VVSVYY545 pKa = 10.05IDD547 pKa = 3.53EE548 pKa = 4.45AHH550 pKa = 7.09DD551 pKa = 4.35FINGLPGDD559 pKa = 4.03LSEE562 pKa = 5.58ALAQARR568 pKa = 11.84SLGGALHH575 pKa = 7.28LAHH578 pKa = 7.04QYY580 pKa = 10.03RR581 pKa = 11.84AQLSPAMVQALEE593 pKa = 4.04TNARR597 pKa = 11.84NKK599 pKa = 10.16IYY601 pKa = 10.46FGLSGTDD608 pKa = 3.21AAAVAHH614 pKa = 6.72LAPEE618 pKa = 4.93LEE620 pKa = 4.22AQDD623 pKa = 5.27FIGLPAYY630 pKa = 9.7HH631 pKa = 7.45AYY633 pKa = 8.04ATLMQQGRR641 pKa = 11.84STGWLSVTTRR651 pKa = 11.84PAPPARR657 pKa = 11.84QEE659 pKa = 3.97PASLYY664 pKa = 9.99AASHH668 pKa = 4.74QRR670 pKa = 11.84YY671 pKa = 8.3GVPATRR677 pKa = 11.84TEE679 pKa = 3.89QQLINLTKK687 pKa = 9.45PSHH690 pKa = 6.69HH691 pKa = 6.54ATQPPDD697 pKa = 3.71SPAKK701 pKa = 10.11PSTGSAAPDD710 pKa = 3.26QPGPIGRR717 pKa = 11.84RR718 pKa = 11.84RR719 pKa = 11.84RR720 pKa = 3.46
MM1 pKa = 7.34NGTWRR6 pKa = 11.84WQEE9 pKa = 3.99LAWDD13 pKa = 4.19SPATPAVGAAALEE26 pKa = 4.17RR27 pKa = 11.84LTTAVEE33 pKa = 3.99VGPVVLEE40 pKa = 3.71LRR42 pKa = 11.84AWAGGAYY49 pKa = 8.59WLAGCQPGRR58 pKa = 11.84AVEE61 pKa = 4.28LKK63 pKa = 10.46QLLGHH68 pKa = 6.71HH69 pKa = 6.6LPARR73 pKa = 11.84LHH75 pKa = 5.93RR76 pKa = 11.84PQRR79 pKa = 11.84QRR81 pKa = 11.84LAVGQAVRR89 pKa = 11.84LQVRR93 pKa = 11.84GNHH96 pKa = 5.39VSTSADD102 pKa = 2.7RR103 pKa = 11.84VVAATRR109 pKa = 11.84ALYY112 pKa = 10.42ACLSGLHH119 pKa = 6.43EE120 pKa = 4.39NQQAVVQLLIGRR132 pKa = 11.84RR133 pKa = 11.84LPSSSFTGPPAPGWRR148 pKa = 11.84EE149 pKa = 3.79LFLGQPAPRR158 pKa = 11.84SPEE161 pKa = 3.72RR162 pKa = 11.84PTRR165 pKa = 11.84TTSEE169 pKa = 3.7QHH171 pKa = 5.69GAAACLRR178 pKa = 11.84LGHH181 pKa = 6.72TGTPAQAHH189 pKa = 4.75QLFSRR194 pKa = 11.84LLGALRR200 pKa = 11.84IVEE203 pKa = 4.26TPRR206 pKa = 11.84ARR208 pKa = 11.84FRR210 pKa = 11.84LSAEE214 pKa = 3.8APDD217 pKa = 3.51RR218 pKa = 11.84LNRR221 pKa = 11.84VSRR224 pKa = 11.84PWAWPLRR231 pKa = 11.84LRR233 pKa = 11.84SGQLTALCGWPIGEE247 pKa = 4.84PPLPLLGDD255 pKa = 3.54LHH257 pKa = 6.83PRR259 pKa = 11.84LLPPPASLVRR269 pKa = 11.84SEE271 pKa = 4.48RR272 pKa = 11.84VIGLASAPGCRR283 pKa = 11.84QPVAIPIRR291 pKa = 11.84DD292 pKa = 3.75AVFHH296 pKa = 5.55THH298 pKa = 7.12LLGPTGTGKK307 pKa = 8.49STVLLSLALADD318 pKa = 4.13VEE320 pKa = 4.12AHH322 pKa = 6.42RR323 pKa = 11.84AVLLLDD329 pKa = 3.97PKK331 pKa = 11.28GDD333 pKa = 3.71LATDD337 pKa = 3.93LLARR341 pKa = 11.84VPEE344 pKa = 4.47GRR346 pKa = 11.84HH347 pKa = 5.18EE348 pKa = 4.35DD349 pKa = 3.68VVVIDD354 pKa = 4.25PTNPAPVGLNPLAGPARR371 pKa = 11.84LAPVTADD378 pKa = 3.31SLLGTLAALFHH389 pKa = 5.8EE390 pKa = 4.71HH391 pKa = 6.02WGVRR395 pKa = 11.84TADD398 pKa = 3.6VLNAALLTLARR409 pKa = 11.84TPGANLLWLPPLLTDD424 pKa = 3.41PAFRR428 pKa = 11.84RR429 pKa = 11.84RR430 pKa = 11.84VLDD433 pKa = 4.38DD434 pKa = 3.31QDD436 pKa = 5.23DD437 pKa = 4.09PLGTGAFWAAYY448 pKa = 6.69EE449 pKa = 4.15AKK451 pKa = 10.14RR452 pKa = 11.84PQAQAVEE459 pKa = 4.14IAPVLNKK466 pKa = 9.82LRR468 pKa = 11.84QLILRR473 pKa = 11.84PHH475 pKa = 6.2LRR477 pKa = 11.84AVLGQSAPRR486 pKa = 11.84FTVEE490 pKa = 3.89EE491 pKa = 4.29LFTRR495 pKa = 11.84KK496 pKa = 9.96RR497 pKa = 11.84IVVVNLNRR505 pKa = 11.84GLLGAEE511 pKa = 4.34AARR514 pKa = 11.84LLGSLLVGQLWSRR527 pKa = 11.84LLARR531 pKa = 11.84QALPPEE537 pKa = 4.28RR538 pKa = 11.84RR539 pKa = 11.84HH540 pKa = 5.23VVSVYY545 pKa = 10.05IDD547 pKa = 3.53EE548 pKa = 4.45AHH550 pKa = 7.09DD551 pKa = 4.35FINGLPGDD559 pKa = 4.03LSEE562 pKa = 5.58ALAQARR568 pKa = 11.84SLGGALHH575 pKa = 7.28LAHH578 pKa = 7.04QYY580 pKa = 10.03RR581 pKa = 11.84AQLSPAMVQALEE593 pKa = 4.04TNARR597 pKa = 11.84NKK599 pKa = 10.16IYY601 pKa = 10.46FGLSGTDD608 pKa = 3.21AAAVAHH614 pKa = 6.72LAPEE618 pKa = 4.93LEE620 pKa = 4.22AQDD623 pKa = 5.27FIGLPAYY630 pKa = 9.7HH631 pKa = 7.45AYY633 pKa = 8.04ATLMQQGRR641 pKa = 11.84STGWLSVTTRR651 pKa = 11.84PAPPARR657 pKa = 11.84QEE659 pKa = 3.97PASLYY664 pKa = 9.99AASHH668 pKa = 4.74QRR670 pKa = 11.84YY671 pKa = 8.3GVPATRR677 pKa = 11.84TEE679 pKa = 3.89QQLINLTKK687 pKa = 9.45PSHH690 pKa = 6.69HH691 pKa = 6.54ATQPPDD697 pKa = 3.71SPAKK701 pKa = 10.11PSTGSAAPDD710 pKa = 3.26QPGPIGRR717 pKa = 11.84RR718 pKa = 11.84RR719 pKa = 11.84RR720 pKa = 3.46
Molecular weight: 77.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
276238 |
37 |
1281 |
286.9 |
31.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.471 ± 0.094 | 0.797 ± 0.023 |
5.829 ± 0.059 | 5.767 ± 0.073 |
3.61 ± 0.05 | 6.691 ± 0.08 |
2.084 ± 0.038 | 6.667 ± 0.069 |
5.664 ± 0.08 | 9.022 ± 0.085 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.32 ± 0.034 | 4.166 ± 0.059 |
3.867 ± 0.059 | 3.864 ± 0.048 |
5.984 ± 0.069 | 6.373 ± 0.076 |
6.019 ± 0.064 | 7.338 ± 0.084 |
1.063 ± 0.031 | 3.402 ± 0.044 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
