
Capybara microvirus Cap3_SP_414
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
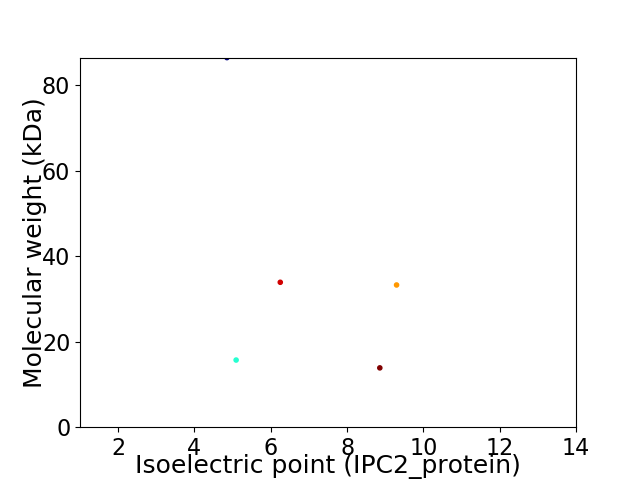
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5U7|A0A4P8W5U7_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_414 OX=2585447 PE=4 SV=1
MM1 pKa = 7.56SNNKK5 pKa = 8.81IFQAEE10 pKa = 4.13IVEE13 pKa = 4.48KK14 pKa = 10.11EE15 pKa = 4.01YY16 pKa = 10.94QSRR19 pKa = 11.84NEE21 pKa = 3.76FDD23 pKa = 3.51FTHH26 pKa = 7.52DD27 pKa = 3.55NLTTLDD33 pKa = 3.7MGNIVPLDD41 pKa = 3.79SVLLFPGDD49 pKa = 3.15ICDD52 pKa = 4.0ISSEE56 pKa = 4.88TYY58 pKa = 10.15FQTHH62 pKa = 6.0PCITNLLSKK71 pKa = 9.51VYY73 pKa = 10.31CDD75 pKa = 3.93EE76 pKa = 5.07LDD78 pKa = 4.76FIAPLSRR85 pKa = 11.84VWTLWQDD92 pKa = 3.71YY93 pKa = 9.94ISPGNGTRR101 pKa = 11.84PLSHH105 pKa = 7.12TDD107 pKa = 3.23PYY109 pKa = 10.12RR110 pKa = 11.84TPTKK114 pKa = 9.88PYY116 pKa = 10.23FVFEE120 pKa = 4.9DD121 pKa = 4.05IILASLQTSFSILNFDD137 pKa = 4.56INFTYY142 pKa = 10.95APMKK146 pKa = 9.89QRR148 pKa = 11.84QFMLIDD154 pKa = 3.57YY155 pKa = 8.78LHH157 pKa = 6.59YY158 pKa = 10.12PIQNFISRR166 pKa = 11.84SDD168 pKa = 3.64LDD170 pKa = 3.91LSNLHH175 pKa = 5.92TFRR178 pKa = 11.84NYY180 pKa = 10.11VKK182 pKa = 10.74LSQTNQTPINLKK194 pKa = 10.54RR195 pKa = 11.84IGLTPNYY202 pKa = 9.61YY203 pKa = 10.15RR204 pKa = 11.84GVANLLNDD212 pKa = 4.89LLPTSLQNKK221 pKa = 9.18IYY223 pKa = 9.32IHH225 pKa = 5.13QVYY228 pKa = 9.86EE229 pKa = 4.21EE230 pKa = 4.31VGKK233 pKa = 10.94VPYY236 pKa = 10.09NAIIYY241 pKa = 10.4LSDD244 pKa = 4.25VINAWLYY251 pKa = 10.25DD252 pKa = 3.72LEE254 pKa = 5.98IIFPVNYY261 pKa = 9.59YY262 pKa = 10.75NDD264 pKa = 3.57VSSVPDD270 pKa = 3.87FNEE273 pKa = 4.35IILLSIIEE281 pKa = 4.33CLANQKK287 pKa = 10.26SDD289 pKa = 3.41PLVYY293 pKa = 10.33CNSDD297 pKa = 2.76IYY299 pKa = 11.41DD300 pKa = 3.99PYY302 pKa = 11.49HH303 pKa = 6.05DD304 pKa = 4.41VKK306 pKa = 11.44YY307 pKa = 10.99NDD309 pKa = 3.09YY310 pKa = 11.3SYY312 pKa = 11.41FRR314 pKa = 11.84NNNLNLVMYY323 pKa = 10.04YY324 pKa = 10.65CDD326 pKa = 3.9FANSFPSEE334 pKa = 4.0TKK336 pKa = 10.44EE337 pKa = 3.65ILKK340 pKa = 10.3KK341 pKa = 10.42CFYY344 pKa = 10.03VAGGVRR350 pKa = 11.84QTIYY354 pKa = 10.52KK355 pKa = 9.84VEE357 pKa = 4.22PNEE360 pKa = 4.85LDD362 pKa = 3.27TTNYY366 pKa = 9.91QVIDD370 pKa = 3.63ALEE373 pKa = 4.41FYY375 pKa = 10.57AYY377 pKa = 10.82LKK379 pKa = 10.04VCCDD383 pKa = 3.37YY384 pKa = 11.06FRR386 pKa = 11.84DD387 pKa = 3.62QNYY390 pKa = 10.58DD391 pKa = 3.04ILEE394 pKa = 4.13VLISEE399 pKa = 4.59SVKK402 pKa = 11.09SNLNSKK408 pKa = 9.84FQLTQEE414 pKa = 5.16GYY416 pKa = 10.26LSPSSIPEE424 pKa = 4.2LFVSCYY430 pKa = 9.97KK431 pKa = 10.31KK432 pKa = 10.72DD433 pKa = 3.77YY434 pKa = 8.15FTTATEE440 pKa = 4.05TAQRR444 pKa = 11.84GNPIEE449 pKa = 4.61IPTTYY454 pKa = 8.77ITLPDD459 pKa = 3.89PVIGGTASKK468 pKa = 11.06VRR470 pKa = 11.84MNVFDD475 pKa = 4.03PQNPFVIFDD484 pKa = 3.93NDD486 pKa = 3.21STMKK490 pKa = 10.4SLSISGTSQPLTVLDD505 pKa = 3.68LRR507 pKa = 11.84KK508 pKa = 10.15YY509 pKa = 10.4ISINHH514 pKa = 5.27YY515 pKa = 10.57VEE517 pKa = 4.68SCLTSGMRR525 pKa = 11.84YY526 pKa = 9.82AEE528 pKa = 4.32YY529 pKa = 9.81FYY531 pKa = 9.87MQWDD535 pKa = 4.07VVVPDD540 pKa = 4.44AVTQEE545 pKa = 4.29TEE547 pKa = 4.58LINAKK552 pKa = 9.3NQIFNFQKK560 pKa = 10.61VVNTGNNDD568 pKa = 3.35LPLGSLASNGFSIDD582 pKa = 3.15SSSSQRR588 pKa = 11.84IHH590 pKa = 6.07QNEE593 pKa = 3.73HH594 pKa = 5.1AFYY597 pKa = 10.25FKK599 pKa = 10.56FIRR602 pKa = 11.84ILPDD606 pKa = 3.41LYY608 pKa = 11.0YY609 pKa = 11.03YY610 pKa = 11.14GGIQRR615 pKa = 11.84NKK617 pKa = 10.35LFQKK621 pKa = 10.61YY622 pKa = 9.91LDD624 pKa = 3.78YY625 pKa = 10.81PLPQFSSLGNEE636 pKa = 3.88EE637 pKa = 4.36IYY639 pKa = 11.19SNEE642 pKa = 3.69VLFQFSNAKK651 pKa = 10.47NIFGYY656 pKa = 9.81NSRR659 pKa = 11.84YY660 pKa = 9.77SYY662 pKa = 11.76SKK664 pKa = 10.76FEE666 pKa = 4.37PNKK669 pKa = 10.28VSGDD673 pKa = 3.63FCNSLSSQAFIRR685 pKa = 11.84NLNEE689 pKa = 3.62EE690 pKa = 4.57DD691 pKa = 3.47SDD693 pKa = 4.27LSLRR697 pKa = 11.84CSYY700 pKa = 10.61EE701 pKa = 3.77FNLVVNDD708 pKa = 3.6KK709 pKa = 11.06RR710 pKa = 11.84PFAVISPNYY719 pKa = 9.88DD720 pKa = 3.0SYY722 pKa = 11.56ICHH725 pKa = 6.8SLIKK729 pKa = 10.16FNNWRR734 pKa = 11.84KK735 pKa = 9.69LPVYY739 pKa = 8.98GLPGIHH745 pKa = 6.98TII747 pKa = 4.58
MM1 pKa = 7.56SNNKK5 pKa = 8.81IFQAEE10 pKa = 4.13IVEE13 pKa = 4.48KK14 pKa = 10.11EE15 pKa = 4.01YY16 pKa = 10.94QSRR19 pKa = 11.84NEE21 pKa = 3.76FDD23 pKa = 3.51FTHH26 pKa = 7.52DD27 pKa = 3.55NLTTLDD33 pKa = 3.7MGNIVPLDD41 pKa = 3.79SVLLFPGDD49 pKa = 3.15ICDD52 pKa = 4.0ISSEE56 pKa = 4.88TYY58 pKa = 10.15FQTHH62 pKa = 6.0PCITNLLSKK71 pKa = 9.51VYY73 pKa = 10.31CDD75 pKa = 3.93EE76 pKa = 5.07LDD78 pKa = 4.76FIAPLSRR85 pKa = 11.84VWTLWQDD92 pKa = 3.71YY93 pKa = 9.94ISPGNGTRR101 pKa = 11.84PLSHH105 pKa = 7.12TDD107 pKa = 3.23PYY109 pKa = 10.12RR110 pKa = 11.84TPTKK114 pKa = 9.88PYY116 pKa = 10.23FVFEE120 pKa = 4.9DD121 pKa = 4.05IILASLQTSFSILNFDD137 pKa = 4.56INFTYY142 pKa = 10.95APMKK146 pKa = 9.89QRR148 pKa = 11.84QFMLIDD154 pKa = 3.57YY155 pKa = 8.78LHH157 pKa = 6.59YY158 pKa = 10.12PIQNFISRR166 pKa = 11.84SDD168 pKa = 3.64LDD170 pKa = 3.91LSNLHH175 pKa = 5.92TFRR178 pKa = 11.84NYY180 pKa = 10.11VKK182 pKa = 10.74LSQTNQTPINLKK194 pKa = 10.54RR195 pKa = 11.84IGLTPNYY202 pKa = 9.61YY203 pKa = 10.15RR204 pKa = 11.84GVANLLNDD212 pKa = 4.89LLPTSLQNKK221 pKa = 9.18IYY223 pKa = 9.32IHH225 pKa = 5.13QVYY228 pKa = 9.86EE229 pKa = 4.21EE230 pKa = 4.31VGKK233 pKa = 10.94VPYY236 pKa = 10.09NAIIYY241 pKa = 10.4LSDD244 pKa = 4.25VINAWLYY251 pKa = 10.25DD252 pKa = 3.72LEE254 pKa = 5.98IIFPVNYY261 pKa = 9.59YY262 pKa = 10.75NDD264 pKa = 3.57VSSVPDD270 pKa = 3.87FNEE273 pKa = 4.35IILLSIIEE281 pKa = 4.33CLANQKK287 pKa = 10.26SDD289 pKa = 3.41PLVYY293 pKa = 10.33CNSDD297 pKa = 2.76IYY299 pKa = 11.41DD300 pKa = 3.99PYY302 pKa = 11.49HH303 pKa = 6.05DD304 pKa = 4.41VKK306 pKa = 11.44YY307 pKa = 10.99NDD309 pKa = 3.09YY310 pKa = 11.3SYY312 pKa = 11.41FRR314 pKa = 11.84NNNLNLVMYY323 pKa = 10.04YY324 pKa = 10.65CDD326 pKa = 3.9FANSFPSEE334 pKa = 4.0TKK336 pKa = 10.44EE337 pKa = 3.65ILKK340 pKa = 10.3KK341 pKa = 10.42CFYY344 pKa = 10.03VAGGVRR350 pKa = 11.84QTIYY354 pKa = 10.52KK355 pKa = 9.84VEE357 pKa = 4.22PNEE360 pKa = 4.85LDD362 pKa = 3.27TTNYY366 pKa = 9.91QVIDD370 pKa = 3.63ALEE373 pKa = 4.41FYY375 pKa = 10.57AYY377 pKa = 10.82LKK379 pKa = 10.04VCCDD383 pKa = 3.37YY384 pKa = 11.06FRR386 pKa = 11.84DD387 pKa = 3.62QNYY390 pKa = 10.58DD391 pKa = 3.04ILEE394 pKa = 4.13VLISEE399 pKa = 4.59SVKK402 pKa = 11.09SNLNSKK408 pKa = 9.84FQLTQEE414 pKa = 5.16GYY416 pKa = 10.26LSPSSIPEE424 pKa = 4.2LFVSCYY430 pKa = 9.97KK431 pKa = 10.31KK432 pKa = 10.72DD433 pKa = 3.77YY434 pKa = 8.15FTTATEE440 pKa = 4.05TAQRR444 pKa = 11.84GNPIEE449 pKa = 4.61IPTTYY454 pKa = 8.77ITLPDD459 pKa = 3.89PVIGGTASKK468 pKa = 11.06VRR470 pKa = 11.84MNVFDD475 pKa = 4.03PQNPFVIFDD484 pKa = 3.93NDD486 pKa = 3.21STMKK490 pKa = 10.4SLSISGTSQPLTVLDD505 pKa = 3.68LRR507 pKa = 11.84KK508 pKa = 10.15YY509 pKa = 10.4ISINHH514 pKa = 5.27YY515 pKa = 10.57VEE517 pKa = 4.68SCLTSGMRR525 pKa = 11.84YY526 pKa = 9.82AEE528 pKa = 4.32YY529 pKa = 9.81FYY531 pKa = 9.87MQWDD535 pKa = 4.07VVVPDD540 pKa = 4.44AVTQEE545 pKa = 4.29TEE547 pKa = 4.58LINAKK552 pKa = 9.3NQIFNFQKK560 pKa = 10.61VVNTGNNDD568 pKa = 3.35LPLGSLASNGFSIDD582 pKa = 3.15SSSSQRR588 pKa = 11.84IHH590 pKa = 6.07QNEE593 pKa = 3.73HH594 pKa = 5.1AFYY597 pKa = 10.25FKK599 pKa = 10.56FIRR602 pKa = 11.84ILPDD606 pKa = 3.41LYY608 pKa = 11.0YY609 pKa = 11.03YY610 pKa = 11.14GGIQRR615 pKa = 11.84NKK617 pKa = 10.35LFQKK621 pKa = 10.61YY622 pKa = 9.91LDD624 pKa = 3.78YY625 pKa = 10.81PLPQFSSLGNEE636 pKa = 3.88EE637 pKa = 4.36IYY639 pKa = 11.19SNEE642 pKa = 3.69VLFQFSNAKK651 pKa = 10.47NIFGYY656 pKa = 9.81NSRR659 pKa = 11.84YY660 pKa = 9.77SYY662 pKa = 11.76SKK664 pKa = 10.76FEE666 pKa = 4.37PNKK669 pKa = 10.28VSGDD673 pKa = 3.63FCNSLSSQAFIRR685 pKa = 11.84NLNEE689 pKa = 3.62EE690 pKa = 4.57DD691 pKa = 3.47SDD693 pKa = 4.27LSLRR697 pKa = 11.84CSYY700 pKa = 10.61EE701 pKa = 3.77FNLVVNDD708 pKa = 3.6KK709 pKa = 11.06RR710 pKa = 11.84PFAVISPNYY719 pKa = 9.88DD720 pKa = 3.0SYY722 pKa = 11.56ICHH725 pKa = 6.8SLIKK729 pKa = 10.16FNNWRR734 pKa = 11.84KK735 pKa = 9.69LPVYY739 pKa = 8.98GLPGIHH745 pKa = 6.98TII747 pKa = 4.58
Molecular weight: 86.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W619|A0A4P8W619_9VIRU Replication initiator protein OS=Capybara microvirus Cap3_SP_414 OX=2585447 PE=4 SV=1
MM1 pKa = 7.56NYY3 pKa = 10.07KK4 pKa = 10.17KK5 pKa = 10.27QFKK8 pKa = 10.25NFRR11 pKa = 11.84NNDD14 pKa = 2.9FFINYY19 pKa = 8.15DD20 pKa = 3.25KK21 pKa = 10.96SYY23 pKa = 11.25KK24 pKa = 9.98NLPTIQGDD32 pKa = 3.86VNRR35 pKa = 11.84GISNLIQRR43 pKa = 11.84YY44 pKa = 8.67NRR46 pKa = 11.84GLPLPNDD53 pKa = 3.48AVQVSFGYY61 pKa = 8.65TLEE64 pKa = 4.35ASSPFTPDD72 pKa = 3.21KK73 pKa = 11.32LEE75 pKa = 4.07LAIKK79 pKa = 10.25SKK81 pKa = 10.67QFNNTLSDD89 pKa = 3.29VKK91 pKa = 10.13TQLEE95 pKa = 4.25KK96 pKa = 10.77QIKK99 pKa = 8.12TKK101 pKa = 9.63EE102 pKa = 4.0LEE104 pKa = 3.92KK105 pKa = 10.74RR106 pKa = 11.84SAVEE110 pKa = 3.48NTGNIEE116 pKa = 4.09TSTTSS121 pKa = 2.78
MM1 pKa = 7.56NYY3 pKa = 10.07KK4 pKa = 10.17KK5 pKa = 10.27QFKK8 pKa = 10.25NFRR11 pKa = 11.84NNDD14 pKa = 2.9FFINYY19 pKa = 8.15DD20 pKa = 3.25KK21 pKa = 10.96SYY23 pKa = 11.25KK24 pKa = 9.98NLPTIQGDD32 pKa = 3.86VNRR35 pKa = 11.84GISNLIQRR43 pKa = 11.84YY44 pKa = 8.67NRR46 pKa = 11.84GLPLPNDD53 pKa = 3.48AVQVSFGYY61 pKa = 8.65TLEE64 pKa = 4.35ASSPFTPDD72 pKa = 3.21KK73 pKa = 11.32LEE75 pKa = 4.07LAIKK79 pKa = 10.25SKK81 pKa = 10.67QFNNTLSDD89 pKa = 3.29VKK91 pKa = 10.13TQLEE95 pKa = 4.25KK96 pKa = 10.77QIKK99 pKa = 8.12TKK101 pKa = 9.63EE102 pKa = 4.0LEE104 pKa = 3.92KK105 pKa = 10.74RR106 pKa = 11.84SAVEE110 pKa = 3.48NTGNIEE116 pKa = 4.09TSTTSS121 pKa = 2.78
Molecular weight: 13.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1574 |
121 |
747 |
314.8 |
36.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.066 ± 1.042 | 1.271 ± 0.416 |
5.972 ± 0.631 | 4.956 ± 0.451 |
6.099 ± 0.907 | 3.177 ± 0.282 |
1.398 ± 0.368 | 7.243 ± 0.554 |
7.306 ± 1.575 | 9.466 ± 0.327 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.334 ± 0.336 | 9.657 ± 0.659 |
4.257 ± 0.704 | 5.083 ± 0.554 |
3.367 ± 0.235 | 7.687 ± 0.791 |
5.337 ± 0.504 | 4.701 ± 0.753 |
0.508 ± 0.129 | 7.116 ± 0.844 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
