
Circo-like virus-Brazil hs1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses; Circo-like virus-Brazil
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
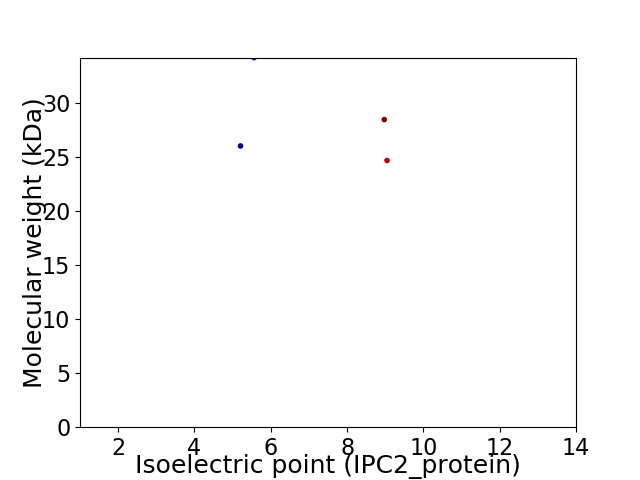
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
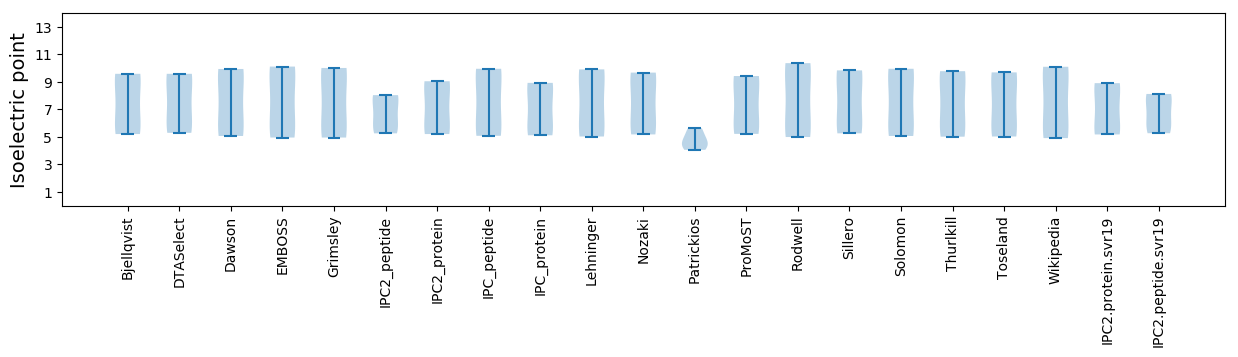
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U3LYW0|U3LYW0_9VIRU Uncharacterized protein OS=Circo-like virus-Brazil hs1 OX=1346815 PE=4 SV=1
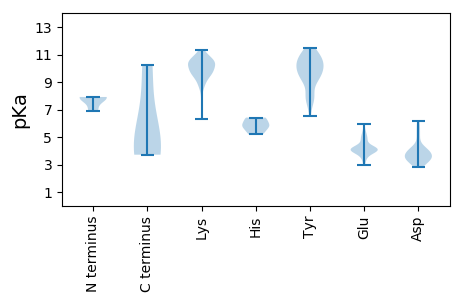
MM1 pKa = 7.89RR2 pKa = 11.84KK3 pKa = 9.85YY4 pKa = 10.67KK5 pKa = 8.87ITKK8 pKa = 9.81NGLIDD13 pKa = 3.92YY14 pKa = 7.43STQKK18 pKa = 10.13PVSTIYY24 pKa = 10.33VGKK27 pKa = 10.2EE28 pKa = 3.25KK29 pKa = 10.62DD30 pKa = 3.45VQTQEE35 pKa = 3.88DD36 pKa = 5.32YY37 pKa = 10.66IYY39 pKa = 10.99KK40 pKa = 8.9DD41 pKa = 2.89TWSLQEE47 pKa = 4.81KK48 pKa = 9.71KK49 pKa = 10.85DD50 pKa = 4.19LMEE53 pKa = 4.32QKK55 pKa = 10.36KK56 pKa = 10.55DD57 pKa = 3.33WIVPEE62 pKa = 4.82FIWKK66 pKa = 9.7EE67 pKa = 3.58EE68 pKa = 3.75WVRR71 pKa = 11.84TGKK74 pKa = 9.99RR75 pKa = 11.84WNTVKK80 pKa = 10.36KK81 pKa = 10.52RR82 pKa = 11.84SLEE85 pKa = 4.07MVNSLNLVNMMIEE98 pKa = 4.01EE99 pKa = 4.06ARR101 pKa = 11.84RR102 pKa = 11.84NVWKK106 pKa = 10.45KK107 pKa = 10.42SKK109 pKa = 10.59KK110 pKa = 8.65EE111 pKa = 4.07SEE113 pKa = 4.48KK114 pKa = 9.7EE115 pKa = 4.34TQNLTSQKK123 pKa = 8.31NTLGFGVDD131 pKa = 3.6TTKK134 pKa = 10.94HH135 pKa = 5.26LVNTEE140 pKa = 2.99QWYY143 pKa = 10.05NLEE146 pKa = 3.92EE147 pKa = 4.1TLRR150 pKa = 11.84PLSEE154 pKa = 5.4FIGEE158 pKa = 4.12DD159 pKa = 3.23QEE161 pKa = 5.37LVNQEE166 pKa = 4.24EE167 pKa = 5.02LPTNVDD173 pKa = 2.92QMHH176 pKa = 6.4TEE178 pKa = 4.02NRR180 pKa = 11.84WEE182 pKa = 4.07NGGMDD187 pKa = 3.43ITEE190 pKa = 4.65HH191 pKa = 5.72LTSLSMTSMDD201 pKa = 4.7GSDD204 pKa = 3.09LMNYY208 pKa = 9.47FDD210 pKa = 6.14ALIGTNTVFQQ220 pKa = 4.07
MM1 pKa = 7.89RR2 pKa = 11.84KK3 pKa = 9.85YY4 pKa = 10.67KK5 pKa = 8.87ITKK8 pKa = 9.81NGLIDD13 pKa = 3.92YY14 pKa = 7.43STQKK18 pKa = 10.13PVSTIYY24 pKa = 10.33VGKK27 pKa = 10.2EE28 pKa = 3.25KK29 pKa = 10.62DD30 pKa = 3.45VQTQEE35 pKa = 3.88DD36 pKa = 5.32YY37 pKa = 10.66IYY39 pKa = 10.99KK40 pKa = 8.9DD41 pKa = 2.89TWSLQEE47 pKa = 4.81KK48 pKa = 9.71KK49 pKa = 10.85DD50 pKa = 4.19LMEE53 pKa = 4.32QKK55 pKa = 10.36KK56 pKa = 10.55DD57 pKa = 3.33WIVPEE62 pKa = 4.82FIWKK66 pKa = 9.7EE67 pKa = 3.58EE68 pKa = 3.75WVRR71 pKa = 11.84TGKK74 pKa = 9.99RR75 pKa = 11.84WNTVKK80 pKa = 10.36KK81 pKa = 10.52RR82 pKa = 11.84SLEE85 pKa = 4.07MVNSLNLVNMMIEE98 pKa = 4.01EE99 pKa = 4.06ARR101 pKa = 11.84RR102 pKa = 11.84NVWKK106 pKa = 10.45KK107 pKa = 10.42SKK109 pKa = 10.59KK110 pKa = 8.65EE111 pKa = 4.07SEE113 pKa = 4.48KK114 pKa = 9.7EE115 pKa = 4.34TQNLTSQKK123 pKa = 8.31NTLGFGVDD131 pKa = 3.6TTKK134 pKa = 10.94HH135 pKa = 5.26LVNTEE140 pKa = 2.99QWYY143 pKa = 10.05NLEE146 pKa = 3.92EE147 pKa = 4.1TLRR150 pKa = 11.84PLSEE154 pKa = 5.4FIGEE158 pKa = 4.12DD159 pKa = 3.23QEE161 pKa = 5.37LVNQEE166 pKa = 4.24EE167 pKa = 5.02LPTNVDD173 pKa = 2.92QMHH176 pKa = 6.4TEE178 pKa = 4.02NRR180 pKa = 11.84WEE182 pKa = 4.07NGGMDD187 pKa = 3.43ITEE190 pKa = 4.65HH191 pKa = 5.72LTSLSMTSMDD201 pKa = 4.7GSDD204 pKa = 3.09LMNYY208 pKa = 9.47FDD210 pKa = 6.14ALIGTNTVFQQ220 pKa = 4.07
Molecular weight: 26.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U3LZA2|U3LZA2_9VIRU ATP-dependent helicase Rep OS=Circo-like virus-Brazil hs1 OX=1346815 PE=3 SV=1
MM1 pKa = 7.83VMRR4 pKa = 11.84LCGWVILDD12 pKa = 3.97QPPKK16 pKa = 9.51RR17 pKa = 11.84TPDD20 pKa = 3.56LEE22 pKa = 4.08IKK24 pKa = 9.81TNLEE28 pKa = 3.73SFLIMSWWDD37 pKa = 4.22DD38 pKa = 3.37IPDD41 pKa = 3.41EE42 pKa = 4.18SFYY45 pKa = 11.45YY46 pKa = 10.08IDD48 pKa = 3.54TPVFKK53 pKa = 10.38SAYY56 pKa = 6.5EE57 pKa = 4.0TLSDD61 pKa = 3.8RR62 pKa = 11.84EE63 pKa = 4.29AIKK66 pKa = 10.45ISNLVEE72 pKa = 3.91LRR74 pKa = 11.84KK75 pKa = 10.03KK76 pKa = 9.09EE77 pKa = 3.97LKK79 pKa = 10.41KK80 pKa = 10.24LNKK83 pKa = 9.96YY84 pKa = 9.08IASLYY89 pKa = 8.19VYY91 pKa = 9.96KK92 pKa = 10.96YY93 pKa = 10.61KK94 pKa = 10.83LDD96 pKa = 3.88GRR98 pKa = 11.84DD99 pKa = 2.86TIYY102 pKa = 11.1NVRR105 pKa = 11.84NKK107 pKa = 9.33ATEE110 pKa = 3.72VRR112 pKa = 11.84NTIEE116 pKa = 4.97KK117 pKa = 9.69IASNAKK123 pKa = 9.81VKK125 pKa = 9.85WSSKK129 pKa = 7.62GWKK132 pKa = 9.42GWDD135 pKa = 3.17IEE137 pKa = 4.25RR138 pKa = 11.84KK139 pKa = 9.6AFKK142 pKa = 10.17KK143 pKa = 10.39FKK145 pKa = 10.45KK146 pKa = 10.06NYY148 pKa = 7.99PNSEE152 pKa = 3.67TDD154 pKa = 3.05YY155 pKa = 10.96TIIVDD160 pKa = 4.2LMAHH164 pKa = 5.87YY165 pKa = 9.25FIKK168 pKa = 10.21MGEE171 pKa = 4.06NLKK174 pKa = 10.82DD175 pKa = 4.08LIDD178 pKa = 4.04RR179 pKa = 11.84RR180 pKa = 11.84LKK182 pKa = 7.2TTSRR186 pKa = 11.84KK187 pKa = 8.91KK188 pKa = 9.4YY189 pKa = 9.34RR190 pKa = 11.84WVATYY195 pKa = 9.63TPTEE199 pKa = 4.01KK200 pKa = 10.57KK201 pKa = 10.32IFMPYY206 pKa = 10.21
MM1 pKa = 7.83VMRR4 pKa = 11.84LCGWVILDD12 pKa = 3.97QPPKK16 pKa = 9.51RR17 pKa = 11.84TPDD20 pKa = 3.56LEE22 pKa = 4.08IKK24 pKa = 9.81TNLEE28 pKa = 3.73SFLIMSWWDD37 pKa = 4.22DD38 pKa = 3.37IPDD41 pKa = 3.41EE42 pKa = 4.18SFYY45 pKa = 11.45YY46 pKa = 10.08IDD48 pKa = 3.54TPVFKK53 pKa = 10.38SAYY56 pKa = 6.5EE57 pKa = 4.0TLSDD61 pKa = 3.8RR62 pKa = 11.84EE63 pKa = 4.29AIKK66 pKa = 10.45ISNLVEE72 pKa = 3.91LRR74 pKa = 11.84KK75 pKa = 10.03KK76 pKa = 9.09EE77 pKa = 3.97LKK79 pKa = 10.41KK80 pKa = 10.24LNKK83 pKa = 9.96YY84 pKa = 9.08IASLYY89 pKa = 8.19VYY91 pKa = 9.96KK92 pKa = 10.96YY93 pKa = 10.61KK94 pKa = 10.83LDD96 pKa = 3.88GRR98 pKa = 11.84DD99 pKa = 2.86TIYY102 pKa = 11.1NVRR105 pKa = 11.84NKK107 pKa = 9.33ATEE110 pKa = 3.72VRR112 pKa = 11.84NTIEE116 pKa = 4.97KK117 pKa = 9.69IASNAKK123 pKa = 9.81VKK125 pKa = 9.85WSSKK129 pKa = 7.62GWKK132 pKa = 9.42GWDD135 pKa = 3.17IEE137 pKa = 4.25RR138 pKa = 11.84KK139 pKa = 9.6AFKK142 pKa = 10.17KK143 pKa = 10.39FKK145 pKa = 10.45KK146 pKa = 10.06NYY148 pKa = 7.99PNSEE152 pKa = 3.67TDD154 pKa = 3.05YY155 pKa = 10.96TIIVDD160 pKa = 4.2LMAHH164 pKa = 5.87YY165 pKa = 9.25FIKK168 pKa = 10.21MGEE171 pKa = 4.06NLKK174 pKa = 10.82DD175 pKa = 4.08LIDD178 pKa = 4.04RR179 pKa = 11.84RR180 pKa = 11.84LKK182 pKa = 7.2TTSRR186 pKa = 11.84KK187 pKa = 8.91KK188 pKa = 9.4YY189 pKa = 9.34RR190 pKa = 11.84WVATYY195 pKa = 9.63TPTEE199 pKa = 4.01KK200 pKa = 10.57KK201 pKa = 10.32IFMPYY206 pKa = 10.21
Molecular weight: 24.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
956 |
206 |
287 |
239.0 |
28.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.033 ± 0.572 | 1.046 ± 0.576 |
6.276 ± 0.327 | 8.577 ± 1.207 |
3.87 ± 0.541 | 5.439 ± 1.129 |
1.046 ± 0.189 | 6.067 ± 0.614 |
9.519 ± 1.583 | 7.845 ± 0.732 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.824 ± 0.498 | 5.649 ± 0.708 |
3.138 ± 0.434 | 3.243 ± 0.877 |
6.695 ± 1.283 | 5.23 ± 0.303 |
6.276 ± 1.075 | 5.753 ± 0.347 |
2.929 ± 0.387 | 5.544 ± 0.623 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
