
Botryotinia calthae
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Helotiales; Sclerotiniaceae; Botryotinia
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
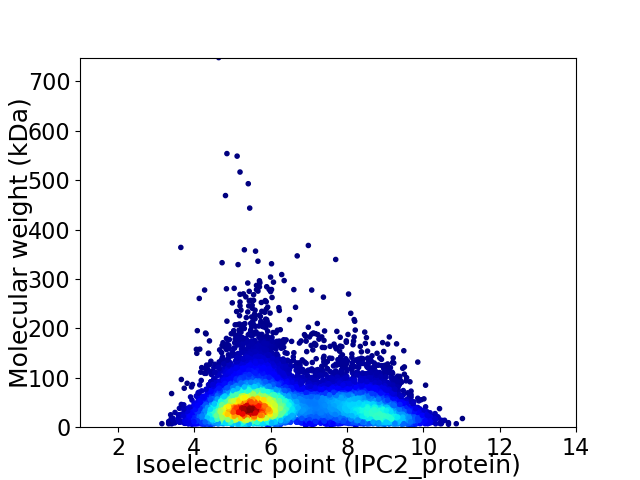
Virtual 2D-PAGE plot for 12263 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
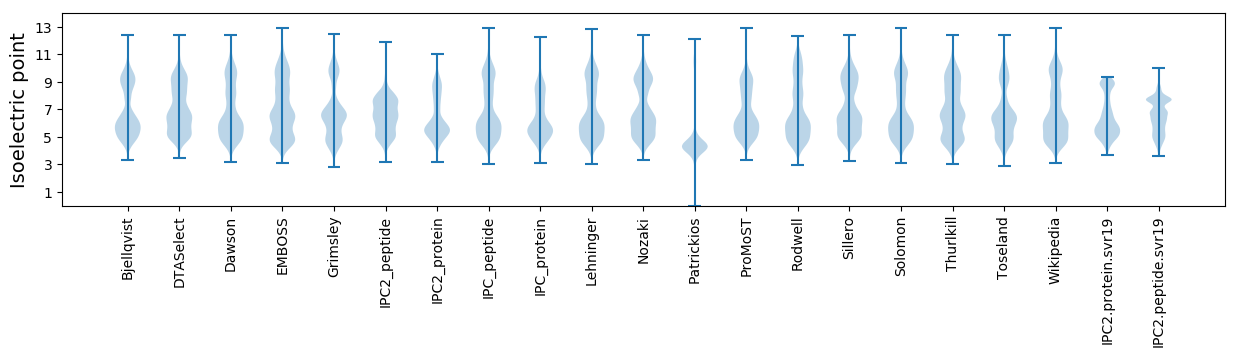
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y8D4R0|A0A4Y8D4R0_9HELO Uncharacterized protein OS=Botryotinia calthae OX=38488 GN=BOTCAL_0125g00260 PE=4 SV=1
MM1 pKa = 7.45SGYY4 pKa = 10.96GNEE7 pKa = 4.28EE8 pKa = 3.66QSDD11 pKa = 4.56SYY13 pKa = 11.54GGQSGNYY20 pKa = 9.08SGKK23 pKa = 9.8QEE25 pKa = 3.89SDD27 pKa = 3.52YY28 pKa = 11.29EE29 pKa = 4.29SRR31 pKa = 11.84DD32 pKa = 3.09NDD34 pKa = 4.12TQSGGYY40 pKa = 9.89GSARR44 pKa = 11.84NNYY47 pKa = 8.41TSGGLGGDD55 pKa = 4.62DD56 pKa = 4.92DD57 pKa = 6.64DD58 pKa = 5.78NDD60 pKa = 3.98NEE62 pKa = 4.62SGDD65 pKa = 4.09AYY67 pKa = 10.74GSTTEE72 pKa = 4.81DD73 pKa = 4.07GYY75 pKa = 11.83GSTRR79 pKa = 11.84KK80 pKa = 10.02DD81 pKa = 3.41DD82 pKa = 3.69TSGGLNNDD90 pKa = 3.78SFSSGDD96 pKa = 3.38TYY98 pKa = 11.58GSTRR102 pKa = 11.84GDD104 pKa = 3.25KK105 pKa = 10.95YY106 pKa = 11.36GSTGKK111 pKa = 10.29DD112 pKa = 2.97DD113 pKa = 3.7TTSGFGQTNTSSEE126 pKa = 4.12DD127 pKa = 3.57AYY129 pKa = 10.91GSTGNNDD136 pKa = 2.79TRR138 pKa = 11.84SGFGQDD144 pKa = 2.94NTTSEE149 pKa = 4.26DD150 pKa = 3.87TYY152 pKa = 11.73GSIGRR157 pKa = 11.84DD158 pKa = 2.97NTTSDD163 pKa = 3.42DD164 pKa = 3.77TYY166 pKa = 11.42SRR168 pKa = 11.84SRR170 pKa = 11.84KK171 pKa = 9.77DD172 pKa = 3.54DD173 pKa = 3.33TSSSYY178 pKa = 11.86NNDD181 pKa = 2.65ATFSGGAFGSTIGAGSDD198 pKa = 3.45SYY200 pKa = 12.18DD201 pKa = 3.34NDD203 pKa = 3.38KK204 pKa = 11.41VGGYY208 pKa = 9.59EE209 pKa = 4.13GVKK212 pKa = 10.29AGSSGYY218 pKa = 10.84GDD220 pKa = 5.48DD221 pKa = 3.65VTGDD225 pKa = 3.54SDD227 pKa = 3.97VSGGYY232 pKa = 10.34GRR234 pKa = 11.84DD235 pKa = 3.67KK236 pKa = 11.22NDD238 pKa = 3.28SQSLGGSGDD247 pKa = 3.52NALSGGYY254 pKa = 10.27GGDD257 pKa = 3.92DD258 pKa = 3.58NQISSGYY265 pKa = 9.88GAGDD269 pKa = 3.44YY270 pKa = 11.05DD271 pKa = 6.26GNDD274 pKa = 3.45QTNSGGYY281 pKa = 9.88GDD283 pKa = 5.3NEE285 pKa = 3.99QSQSGLKK292 pKa = 10.1KK293 pKa = 10.04GLEE296 pKa = 3.99IAKK299 pKa = 9.84EE300 pKa = 4.12LYY302 pKa = 9.93HH303 pKa = 7.12KK304 pKa = 10.81NN305 pKa = 3.23
MM1 pKa = 7.45SGYY4 pKa = 10.96GNEE7 pKa = 4.28EE8 pKa = 3.66QSDD11 pKa = 4.56SYY13 pKa = 11.54GGQSGNYY20 pKa = 9.08SGKK23 pKa = 9.8QEE25 pKa = 3.89SDD27 pKa = 3.52YY28 pKa = 11.29EE29 pKa = 4.29SRR31 pKa = 11.84DD32 pKa = 3.09NDD34 pKa = 4.12TQSGGYY40 pKa = 9.89GSARR44 pKa = 11.84NNYY47 pKa = 8.41TSGGLGGDD55 pKa = 4.62DD56 pKa = 4.92DD57 pKa = 6.64DD58 pKa = 5.78NDD60 pKa = 3.98NEE62 pKa = 4.62SGDD65 pKa = 4.09AYY67 pKa = 10.74GSTTEE72 pKa = 4.81DD73 pKa = 4.07GYY75 pKa = 11.83GSTRR79 pKa = 11.84KK80 pKa = 10.02DD81 pKa = 3.41DD82 pKa = 3.69TSGGLNNDD90 pKa = 3.78SFSSGDD96 pKa = 3.38TYY98 pKa = 11.58GSTRR102 pKa = 11.84GDD104 pKa = 3.25KK105 pKa = 10.95YY106 pKa = 11.36GSTGKK111 pKa = 10.29DD112 pKa = 2.97DD113 pKa = 3.7TTSGFGQTNTSSEE126 pKa = 4.12DD127 pKa = 3.57AYY129 pKa = 10.91GSTGNNDD136 pKa = 2.79TRR138 pKa = 11.84SGFGQDD144 pKa = 2.94NTTSEE149 pKa = 4.26DD150 pKa = 3.87TYY152 pKa = 11.73GSIGRR157 pKa = 11.84DD158 pKa = 2.97NTTSDD163 pKa = 3.42DD164 pKa = 3.77TYY166 pKa = 11.42SRR168 pKa = 11.84SRR170 pKa = 11.84KK171 pKa = 9.77DD172 pKa = 3.54DD173 pKa = 3.33TSSSYY178 pKa = 11.86NNDD181 pKa = 2.65ATFSGGAFGSTIGAGSDD198 pKa = 3.45SYY200 pKa = 12.18DD201 pKa = 3.34NDD203 pKa = 3.38KK204 pKa = 11.41VGGYY208 pKa = 9.59EE209 pKa = 4.13GVKK212 pKa = 10.29AGSSGYY218 pKa = 10.84GDD220 pKa = 5.48DD221 pKa = 3.65VTGDD225 pKa = 3.54SDD227 pKa = 3.97VSGGYY232 pKa = 10.34GRR234 pKa = 11.84DD235 pKa = 3.67KK236 pKa = 11.22NDD238 pKa = 3.28SQSLGGSGDD247 pKa = 3.52NALSGGYY254 pKa = 10.27GGDD257 pKa = 3.92DD258 pKa = 3.58NQISSGYY265 pKa = 9.88GAGDD269 pKa = 3.44YY270 pKa = 11.05DD271 pKa = 6.26GNDD274 pKa = 3.45QTNSGGYY281 pKa = 9.88GDD283 pKa = 5.3NEE285 pKa = 3.99QSQSGLKK292 pKa = 10.1KK293 pKa = 10.04GLEE296 pKa = 3.99IAKK299 pKa = 9.84EE300 pKa = 4.12LYY302 pKa = 9.93HH303 pKa = 7.12KK304 pKa = 10.81NN305 pKa = 3.23
Molecular weight: 31.32 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y8CQS2|A0A4Y8CQS2_9HELO Uncharacterized protein OS=Botryotinia calthae OX=38488 GN=BOTCAL_0447g00050 PE=4 SV=1
MM1 pKa = 7.11ATAEE5 pKa = 4.34PSPASSSNRR14 pKa = 11.84PPRR17 pKa = 11.84TRR19 pKa = 11.84TRR21 pKa = 11.84RR22 pKa = 11.84PRR24 pKa = 11.84RR25 pKa = 11.84GGHH28 pKa = 6.46RR29 pKa = 11.84NTGPAPADD37 pKa = 3.43ANTNATPNQTTQAPNGPLALRR58 pKa = 11.84PVSVASPPGSNPSTPSNPRR77 pKa = 11.84NSNANNRR84 pKa = 11.84GGGNGRR90 pKa = 11.84RR91 pKa = 11.84GGFGRR96 pKa = 11.84GGRR99 pKa = 11.84RR100 pKa = 11.84GGGRR104 pKa = 11.84GQPMANGRR112 pKa = 11.84VFGGQLTSNLHH123 pKa = 5.78TGPTSDD129 pKa = 4.43AGSLAGNAPAFTPGQPIAARR149 pKa = 11.84PGPSRR154 pKa = 11.84APRR157 pKa = 11.84ARR159 pKa = 11.84RR160 pKa = 11.84MSKK163 pKa = 9.68SQAADD168 pKa = 3.17LPTRR172 pKa = 11.84THH174 pKa = 6.58EE175 pKa = 5.44DD176 pKa = 2.94ITNGQYY182 pKa = 10.63EE183 pKa = 4.4CAICTNEE190 pKa = 3.78VLPNSKK196 pKa = 8.8IWNCKK201 pKa = 7.95SCWSVLHH208 pKa = 6.05LSCVKK213 pKa = 10.01KK214 pKa = 9.29WSKK217 pKa = 11.41NEE219 pKa = 3.82VSTHH223 pKa = 3.98QQRR226 pKa = 11.84AVEE229 pKa = 4.27NGEE232 pKa = 4.26LPPPRR237 pKa = 11.84QWRR240 pKa = 11.84CPGCNLPKK248 pKa = 10.16EE249 pKa = 4.57DD250 pKa = 4.92LPISYY255 pKa = 8.33TCWCEE260 pKa = 3.94KK261 pKa = 10.23EE262 pKa = 4.57LGYY265 pKa = 10.65HH266 pKa = 6.73HH267 pKa = 7.14ILVGKK272 pKa = 8.93VVQKK276 pKa = 10.96SGLDD280 pKa = 3.28TVLIHH285 pKa = 7.46AISCAMLDD293 pKa = 3.66RR294 pKa = 11.84VHH296 pKa = 7.06LALIWGLHH304 pKa = 5.21FLASVAKK311 pKa = 10.29RR312 pKa = 11.84RR313 pKa = 11.84FQEE316 pKa = 4.24DD317 pKa = 3.1VWIRR321 pKa = 11.84STKK324 pKa = 10.33ADD326 pKa = 3.34GVVGKK331 pKa = 9.35FAMMYY336 pKa = 9.79FPAEE340 pKa = 3.89KK341 pKa = 9.5TIVSEE346 pKa = 4.26VVMRR350 pKa = 11.84ASEE353 pKa = 4.2AVVKK357 pKa = 10.13FLLNQHH363 pKa = 6.68AIVVKK368 pKa = 10.29SRR370 pKa = 11.84KK371 pKa = 9.5LYY373 pKa = 9.74LARR376 pKa = 11.84NAMKK380 pKa = 10.24SVKK383 pKa = 9.73ARR385 pKa = 11.84HH386 pKa = 4.31QPIRR390 pKa = 11.84LMVKK394 pKa = 8.56VWPRR398 pKa = 11.84EE399 pKa = 3.75HH400 pKa = 6.91GLDD403 pKa = 3.54HH404 pKa = 6.66LTVALNAVDD413 pKa = 4.33PLIAVNRR420 pKa = 11.84IITVRR425 pKa = 11.84LDD427 pKa = 3.36ATLKK431 pKa = 9.62TYY433 pKa = 10.63YY434 pKa = 10.4LHH436 pKa = 6.67TVHH439 pKa = 6.94CRR441 pKa = 11.84QTLSQLVPAEE451 pKa = 3.98KK452 pKa = 10.32RR453 pKa = 11.84QFLHH457 pKa = 6.1SSKK460 pKa = 9.42PHH462 pKa = 5.8EE463 pKa = 5.16PIAPSIFLTVKK474 pKa = 10.36RR475 pKa = 11.84SANCRR480 pKa = 3.45
MM1 pKa = 7.11ATAEE5 pKa = 4.34PSPASSSNRR14 pKa = 11.84PPRR17 pKa = 11.84TRR19 pKa = 11.84TRR21 pKa = 11.84RR22 pKa = 11.84PRR24 pKa = 11.84RR25 pKa = 11.84GGHH28 pKa = 6.46RR29 pKa = 11.84NTGPAPADD37 pKa = 3.43ANTNATPNQTTQAPNGPLALRR58 pKa = 11.84PVSVASPPGSNPSTPSNPRR77 pKa = 11.84NSNANNRR84 pKa = 11.84GGGNGRR90 pKa = 11.84RR91 pKa = 11.84GGFGRR96 pKa = 11.84GGRR99 pKa = 11.84RR100 pKa = 11.84GGGRR104 pKa = 11.84GQPMANGRR112 pKa = 11.84VFGGQLTSNLHH123 pKa = 5.78TGPTSDD129 pKa = 4.43AGSLAGNAPAFTPGQPIAARR149 pKa = 11.84PGPSRR154 pKa = 11.84APRR157 pKa = 11.84ARR159 pKa = 11.84RR160 pKa = 11.84MSKK163 pKa = 9.68SQAADD168 pKa = 3.17LPTRR172 pKa = 11.84THH174 pKa = 6.58EE175 pKa = 5.44DD176 pKa = 2.94ITNGQYY182 pKa = 10.63EE183 pKa = 4.4CAICTNEE190 pKa = 3.78VLPNSKK196 pKa = 8.8IWNCKK201 pKa = 7.95SCWSVLHH208 pKa = 6.05LSCVKK213 pKa = 10.01KK214 pKa = 9.29WSKK217 pKa = 11.41NEE219 pKa = 3.82VSTHH223 pKa = 3.98QQRR226 pKa = 11.84AVEE229 pKa = 4.27NGEE232 pKa = 4.26LPPPRR237 pKa = 11.84QWRR240 pKa = 11.84CPGCNLPKK248 pKa = 10.16EE249 pKa = 4.57DD250 pKa = 4.92LPISYY255 pKa = 8.33TCWCEE260 pKa = 3.94KK261 pKa = 10.23EE262 pKa = 4.57LGYY265 pKa = 10.65HH266 pKa = 6.73HH267 pKa = 7.14ILVGKK272 pKa = 8.93VVQKK276 pKa = 10.96SGLDD280 pKa = 3.28TVLIHH285 pKa = 7.46AISCAMLDD293 pKa = 3.66RR294 pKa = 11.84VHH296 pKa = 7.06LALIWGLHH304 pKa = 5.21FLASVAKK311 pKa = 10.29RR312 pKa = 11.84RR313 pKa = 11.84FQEE316 pKa = 4.24DD317 pKa = 3.1VWIRR321 pKa = 11.84STKK324 pKa = 10.33ADD326 pKa = 3.34GVVGKK331 pKa = 9.35FAMMYY336 pKa = 9.79FPAEE340 pKa = 3.89KK341 pKa = 9.5TIVSEE346 pKa = 4.26VVMRR350 pKa = 11.84ASEE353 pKa = 4.2AVVKK357 pKa = 10.13FLLNQHH363 pKa = 6.68AIVVKK368 pKa = 10.29SRR370 pKa = 11.84KK371 pKa = 9.5LYY373 pKa = 9.74LARR376 pKa = 11.84NAMKK380 pKa = 10.24SVKK383 pKa = 9.73ARR385 pKa = 11.84HH386 pKa = 4.31QPIRR390 pKa = 11.84LMVKK394 pKa = 8.56VWPRR398 pKa = 11.84EE399 pKa = 3.75HH400 pKa = 6.91GLDD403 pKa = 3.54HH404 pKa = 6.66LTVALNAVDD413 pKa = 4.33PLIAVNRR420 pKa = 11.84IITVRR425 pKa = 11.84LDD427 pKa = 3.36ATLKK431 pKa = 9.62TYY433 pKa = 10.63YY434 pKa = 10.4LHH436 pKa = 6.67TVHH439 pKa = 6.94CRR441 pKa = 11.84QTLSQLVPAEE451 pKa = 3.98KK452 pKa = 10.32RR453 pKa = 11.84QFLHH457 pKa = 6.1SSKK460 pKa = 9.42PHH462 pKa = 5.8EE463 pKa = 5.16PIAPSIFLTVKK474 pKa = 10.36RR475 pKa = 11.84SANCRR480 pKa = 3.45
Molecular weight: 52.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5604297 |
21 |
6720 |
457.0 |
50.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.634 ± 0.017 | 1.194 ± 0.009 |
5.559 ± 0.018 | 6.558 ± 0.028 |
3.758 ± 0.012 | 6.715 ± 0.024 |
2.274 ± 0.01 | 5.562 ± 0.015 |
5.481 ± 0.024 | 8.558 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.24 ± 0.008 | 4.296 ± 0.012 |
5.714 ± 0.026 | 3.879 ± 0.017 |
5.617 ± 0.022 | 8.814 ± 0.03 |
6.169 ± 0.019 | 5.754 ± 0.016 |
1.388 ± 0.009 | 2.838 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
