
Acidianus tailed spindle virus
Taxonomy: Viruses; Bicaudaviridae; unclassified Bicaudaviridae
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
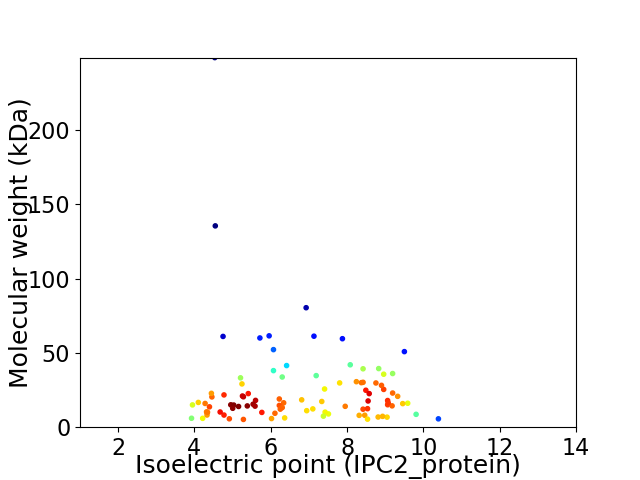
Virtual 2D-PAGE plot for 95 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A125SJA0|A0A125SJA0_9VIRU Uncharacterized protein OS=Acidianus tailed spindle virus OX=1797140 GN=ATSV_C110 PE=4 SV=1
MM1 pKa = 8.13DD2 pKa = 4.18YY3 pKa = 11.31EE4 pKa = 4.76EE5 pKa = 4.82VIQTIKK11 pKa = 11.01EE12 pKa = 4.03FFNSKK17 pKa = 9.51NAEE20 pKa = 4.18FNVGQPQKK28 pKa = 9.85TEE30 pKa = 3.84NNTISIVVDD39 pKa = 4.45FKK41 pKa = 11.69EE42 pKa = 4.58PIEE45 pKa = 4.09LTEE48 pKa = 4.04EE49 pKa = 4.46EE50 pKa = 4.71YY51 pKa = 11.37DD52 pKa = 4.18DD53 pKa = 6.62LYY55 pKa = 11.76DD56 pKa = 4.56FLDD59 pKa = 3.63NLGFYY64 pKa = 9.85PINGAEE70 pKa = 4.01NQVNNYY76 pKa = 9.09IGYY79 pKa = 8.94KK80 pKa = 8.96SWVGVDD86 pKa = 3.3EE87 pKa = 4.99FEE89 pKa = 5.34NDD91 pKa = 3.79DD92 pKa = 3.63GDD94 pKa = 4.1EE95 pKa = 4.37VIVYY99 pKa = 6.76YY100 pKa = 10.4TKK102 pKa = 10.67VCTGGKK108 pKa = 5.06TTFFIKK114 pKa = 10.41KK115 pKa = 9.65IEE117 pKa = 4.04VEE119 pKa = 4.43LNIQEE124 pKa = 4.35NTNEE128 pKa = 3.9LL129 pKa = 3.64
MM1 pKa = 8.13DD2 pKa = 4.18YY3 pKa = 11.31EE4 pKa = 4.76EE5 pKa = 4.82VIQTIKK11 pKa = 11.01EE12 pKa = 4.03FFNSKK17 pKa = 9.51NAEE20 pKa = 4.18FNVGQPQKK28 pKa = 9.85TEE30 pKa = 3.84NNTISIVVDD39 pKa = 4.45FKK41 pKa = 11.69EE42 pKa = 4.58PIEE45 pKa = 4.09LTEE48 pKa = 4.04EE49 pKa = 4.46EE50 pKa = 4.71YY51 pKa = 11.37DD52 pKa = 4.18DD53 pKa = 6.62LYY55 pKa = 11.76DD56 pKa = 4.56FLDD59 pKa = 3.63NLGFYY64 pKa = 9.85PINGAEE70 pKa = 4.01NQVNNYY76 pKa = 9.09IGYY79 pKa = 8.94KK80 pKa = 8.96SWVGVDD86 pKa = 3.3EE87 pKa = 4.99FEE89 pKa = 5.34NDD91 pKa = 3.79DD92 pKa = 3.63GDD94 pKa = 4.1EE95 pKa = 4.37VIVYY99 pKa = 6.76YY100 pKa = 10.4TKK102 pKa = 10.67VCTGGKK108 pKa = 5.06TTFFIKK114 pKa = 10.41KK115 pKa = 9.65IEE117 pKa = 4.04VEE119 pKa = 4.43LNIQEE124 pKa = 4.35NTNEE128 pKa = 3.9LL129 pKa = 3.64
Molecular weight: 15.01 kDa
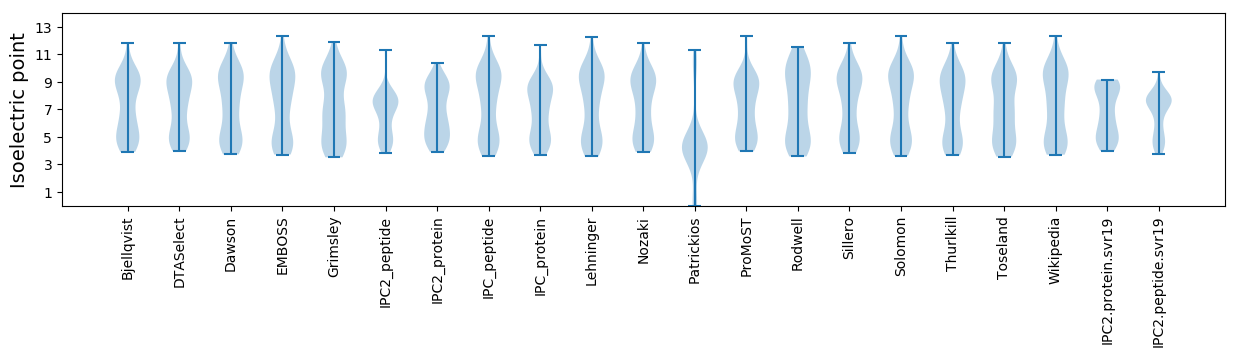
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A125SJ73|A0A125SJ73_9VIRU Transposase OS=Acidianus tailed spindle virus OX=1797140 GN=ATSV_D194 PE=4 SV=1
MM1 pKa = 7.63LKK3 pKa = 10.16NLRR6 pKa = 11.84LNNFQPQEE14 pKa = 3.94EE15 pKa = 4.88YY16 pKa = 11.31VYY18 pKa = 10.04LTYY21 pKa = 10.96SLKK24 pKa = 10.68NNKK27 pKa = 9.32KK28 pKa = 9.75EE29 pKa = 3.86EE30 pKa = 4.19TKK32 pKa = 10.47ILLEE36 pKa = 4.06NYY38 pKa = 9.45RR39 pKa = 11.84VLLQKK44 pKa = 10.95ALDD47 pKa = 3.91WLWEE51 pKa = 3.91RR52 pKa = 11.84TKK54 pKa = 10.44IEE56 pKa = 3.96RR57 pKa = 11.84KK58 pKa = 8.19EE59 pKa = 3.86VKK61 pKa = 9.72KK62 pKa = 10.82GKK64 pKa = 10.03KK65 pKa = 9.5VLTKK69 pKa = 10.47IKK71 pKa = 9.29ITLPKK76 pKa = 9.92KK77 pKa = 9.49KK78 pKa = 9.66EE79 pKa = 3.93VYY81 pKa = 8.41KK82 pKa = 9.68TLRR85 pKa = 11.84DD86 pKa = 3.61EE87 pKa = 4.34LEE89 pKa = 4.81KK90 pKa = 10.68INKK93 pKa = 8.53LASHH97 pKa = 6.23YY98 pKa = 9.95VDD100 pKa = 4.19KK101 pKa = 11.03AISDD105 pKa = 3.9AYY107 pKa = 10.98SILNSWRR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84AEE118 pKa = 3.76KK119 pKa = 10.6GQASLRR125 pKa = 11.84KK126 pKa = 7.41PTLRR130 pKa = 11.84KK131 pKa = 9.31VYY133 pKa = 10.39VRR135 pKa = 11.84VKK137 pKa = 8.49STLRR141 pKa = 11.84KK142 pKa = 9.86VEE144 pKa = 4.12GEE146 pKa = 4.14SVRR149 pKa = 11.84ITVRR153 pKa = 11.84PYY155 pKa = 10.63EE156 pKa = 4.1YY157 pKa = 10.28VTFSWSHH164 pKa = 4.22KK165 pKa = 7.46WFSRR169 pKa = 11.84RR170 pKa = 11.84VEE172 pKa = 3.99GLEE175 pKa = 3.98LGEE178 pKa = 4.16PVIKK182 pKa = 9.84EE183 pKa = 4.33DD184 pKa = 4.34KK185 pKa = 9.11VHH187 pKa = 6.98LPFRR191 pKa = 11.84YY192 pKa = 9.32KK193 pKa = 10.86LPWSTPLDD201 pKa = 3.83FLSIDD206 pKa = 3.76SNLYY210 pKa = 9.01TLDD213 pKa = 4.08AYY215 pKa = 10.67DD216 pKa = 3.83GEE218 pKa = 4.4KK219 pKa = 10.57FITFSMKK226 pKa = 10.05EE227 pKa = 4.0LYY229 pKa = 9.96SLKK232 pKa = 10.43FGMEE236 pKa = 4.69LKK238 pKa = 10.54RR239 pKa = 11.84SRR241 pKa = 11.84IQSFASKK248 pKa = 9.93HH249 pKa = 4.16GRR251 pKa = 11.84KK252 pKa = 9.81GKK254 pKa = 10.36VLMRR258 pKa = 11.84KK259 pKa = 8.65YY260 pKa = 10.38SHH262 pKa = 6.9RR263 pKa = 11.84EE264 pKa = 3.34RR265 pKa = 11.84NRR267 pKa = 11.84VLDD270 pKa = 3.91YY271 pKa = 10.86VHH273 pKa = 6.99KK274 pKa = 10.32FVNKK278 pKa = 9.9LLEE281 pKa = 4.49IYY283 pKa = 9.69PVTIFAVEE291 pKa = 4.11KK292 pKa = 10.65LNKK295 pKa = 9.8QSMFQDD301 pKa = 3.79ANDD304 pKa = 3.95KK305 pKa = 10.71LSKK308 pKa = 10.25KK309 pKa = 9.4VSRR312 pKa = 11.84TVWRR316 pKa = 11.84TIHH319 pKa = 6.99RR320 pKa = 11.84VLKK323 pKa = 10.56YY324 pKa = 9.9KK325 pKa = 10.85APLYY329 pKa = 10.4GSFVKK334 pKa = 10.51EE335 pKa = 3.95VDD337 pKa = 3.07PHH339 pKa = 5.85LTSKK343 pKa = 10.44SCPRR347 pKa = 11.84CGWVSRR353 pKa = 11.84KK354 pKa = 9.12VGRR357 pKa = 11.84TFRR360 pKa = 11.84CEE362 pKa = 3.49RR363 pKa = 11.84CGFTLDD369 pKa = 3.48RR370 pKa = 11.84QLNASLNIYY379 pKa = 10.63LKK381 pKa = 9.76MCGFPHH387 pKa = 7.05ILGIPRR393 pKa = 11.84LWVGVIPLKK402 pKa = 10.52GRR404 pKa = 11.84RR405 pKa = 11.84GNGFPRR411 pKa = 11.84DD412 pKa = 3.65SVEE415 pKa = 4.03AQGLRR420 pKa = 11.84IDD422 pKa = 3.38IKK424 pKa = 10.96YY425 pKa = 10.77YY426 pKa = 10.66EE427 pKa = 4.16ILL429 pKa = 4.02
MM1 pKa = 7.63LKK3 pKa = 10.16NLRR6 pKa = 11.84LNNFQPQEE14 pKa = 3.94EE15 pKa = 4.88YY16 pKa = 11.31VYY18 pKa = 10.04LTYY21 pKa = 10.96SLKK24 pKa = 10.68NNKK27 pKa = 9.32KK28 pKa = 9.75EE29 pKa = 3.86EE30 pKa = 4.19TKK32 pKa = 10.47ILLEE36 pKa = 4.06NYY38 pKa = 9.45RR39 pKa = 11.84VLLQKK44 pKa = 10.95ALDD47 pKa = 3.91WLWEE51 pKa = 3.91RR52 pKa = 11.84TKK54 pKa = 10.44IEE56 pKa = 3.96RR57 pKa = 11.84KK58 pKa = 8.19EE59 pKa = 3.86VKK61 pKa = 9.72KK62 pKa = 10.82GKK64 pKa = 10.03KK65 pKa = 9.5VLTKK69 pKa = 10.47IKK71 pKa = 9.29ITLPKK76 pKa = 9.92KK77 pKa = 9.49KK78 pKa = 9.66EE79 pKa = 3.93VYY81 pKa = 8.41KK82 pKa = 9.68TLRR85 pKa = 11.84DD86 pKa = 3.61EE87 pKa = 4.34LEE89 pKa = 4.81KK90 pKa = 10.68INKK93 pKa = 8.53LASHH97 pKa = 6.23YY98 pKa = 9.95VDD100 pKa = 4.19KK101 pKa = 11.03AISDD105 pKa = 3.9AYY107 pKa = 10.98SILNSWRR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84AEE118 pKa = 3.76KK119 pKa = 10.6GQASLRR125 pKa = 11.84KK126 pKa = 7.41PTLRR130 pKa = 11.84KK131 pKa = 9.31VYY133 pKa = 10.39VRR135 pKa = 11.84VKK137 pKa = 8.49STLRR141 pKa = 11.84KK142 pKa = 9.86VEE144 pKa = 4.12GEE146 pKa = 4.14SVRR149 pKa = 11.84ITVRR153 pKa = 11.84PYY155 pKa = 10.63EE156 pKa = 4.1YY157 pKa = 10.28VTFSWSHH164 pKa = 4.22KK165 pKa = 7.46WFSRR169 pKa = 11.84RR170 pKa = 11.84VEE172 pKa = 3.99GLEE175 pKa = 3.98LGEE178 pKa = 4.16PVIKK182 pKa = 9.84EE183 pKa = 4.33DD184 pKa = 4.34KK185 pKa = 9.11VHH187 pKa = 6.98LPFRR191 pKa = 11.84YY192 pKa = 9.32KK193 pKa = 10.86LPWSTPLDD201 pKa = 3.83FLSIDD206 pKa = 3.76SNLYY210 pKa = 9.01TLDD213 pKa = 4.08AYY215 pKa = 10.67DD216 pKa = 3.83GEE218 pKa = 4.4KK219 pKa = 10.57FITFSMKK226 pKa = 10.05EE227 pKa = 4.0LYY229 pKa = 9.96SLKK232 pKa = 10.43FGMEE236 pKa = 4.69LKK238 pKa = 10.54RR239 pKa = 11.84SRR241 pKa = 11.84IQSFASKK248 pKa = 9.93HH249 pKa = 4.16GRR251 pKa = 11.84KK252 pKa = 9.81GKK254 pKa = 10.36VLMRR258 pKa = 11.84KK259 pKa = 8.65YY260 pKa = 10.38SHH262 pKa = 6.9RR263 pKa = 11.84EE264 pKa = 3.34RR265 pKa = 11.84NRR267 pKa = 11.84VLDD270 pKa = 3.91YY271 pKa = 10.86VHH273 pKa = 6.99KK274 pKa = 10.32FVNKK278 pKa = 9.9LLEE281 pKa = 4.49IYY283 pKa = 9.69PVTIFAVEE291 pKa = 4.11KK292 pKa = 10.65LNKK295 pKa = 9.8QSMFQDD301 pKa = 3.79ANDD304 pKa = 3.95KK305 pKa = 10.71LSKK308 pKa = 10.25KK309 pKa = 9.4VSRR312 pKa = 11.84TVWRR316 pKa = 11.84TIHH319 pKa = 6.99RR320 pKa = 11.84VLKK323 pKa = 10.56YY324 pKa = 9.9KK325 pKa = 10.85APLYY329 pKa = 10.4GSFVKK334 pKa = 10.51EE335 pKa = 3.95VDD337 pKa = 3.07PHH339 pKa = 5.85LTSKK343 pKa = 10.44SCPRR347 pKa = 11.84CGWVSRR353 pKa = 11.84KK354 pKa = 9.12VGRR357 pKa = 11.84TFRR360 pKa = 11.84CEE362 pKa = 3.49RR363 pKa = 11.84CGFTLDD369 pKa = 3.48RR370 pKa = 11.84QLNASLNIYY379 pKa = 10.63LKK381 pKa = 9.76MCGFPHH387 pKa = 7.05ILGIPRR393 pKa = 11.84LWVGVIPLKK402 pKa = 10.52GRR404 pKa = 11.84RR405 pKa = 11.84GNGFPRR411 pKa = 11.84DD412 pKa = 3.65SVEE415 pKa = 4.03AQGLRR420 pKa = 11.84IDD422 pKa = 3.38IKK424 pKa = 10.96YY425 pKa = 10.77YY426 pKa = 10.66EE427 pKa = 4.16ILL429 pKa = 4.02
Molecular weight: 50.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
20891 |
45 |
2246 |
219.9 |
25.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.82 ± 0.249 | 1.063 ± 0.101 |
5.194 ± 0.261 | 6.596 ± 0.418 |
4.471 ± 0.284 | 4.562 ± 0.224 |
1.225 ± 0.115 | 9.296 ± 0.331 |
8.027 ± 0.537 | 9.913 ± 0.327 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.02 ± 0.138 | 6.28 ± 0.314 |
3.695 ± 0.301 | 3.049 ± 0.327 |
3.59 ± 0.39 | 7.467 ± 0.661 |
5.572 ± 0.475 | 6.989 ± 0.27 |
0.723 ± 0.094 | 5.447 ± 0.199 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
